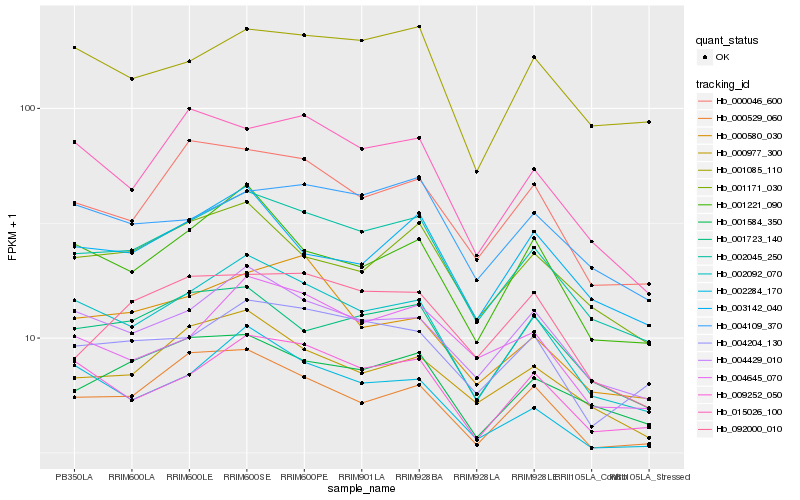
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_250 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002092_070 |
0.0477133454 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 3 |
Hb_004429_010 |
0.0612600961 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_001723_140 |
0.0680500768 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_009252_050 |
0.06875339 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000046_600 |
0.068900137 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 7 |
Hb_000529_060 |
0.0714098165 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001171_030 |
0.0722090643 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 9 |
Hb_004204_130 |
0.0723565928 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 10 |
Hb_000977_300 |
0.07236524 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 11 |
Hb_001584_350 |
0.0742996749 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 12 |
Hb_002284_170 |
0.0745502142 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 13 |
Hb_004645_070 |
0.0756066081 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_004109_370 |
0.0757721251 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 15 |
Hb_092000_010 |
0.0757753698 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 16 |
Hb_001085_110 |
0.077204419 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 17 |
Hb_001221_090 |
0.0783011652 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003142_040 |
0.0813484845 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 19 |
Hb_015026_100 |
0.0816148125 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 20 |
Hb_000580_030 |
0.0817229115 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |