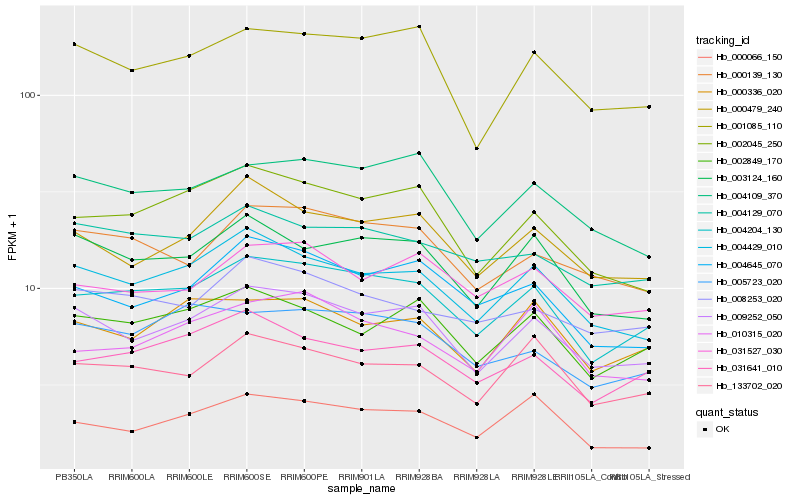
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004204_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 2 |
Hb_031641_010 |
0.062163721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009252_050 |
0.0631382996 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_004429_010 |
0.0707603396 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_002045_250 |
0.0723565928 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 6 |
Hb_002849_170 |
0.073353812 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 7 |
Hb_133702_020 |
0.0754032332 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003124_160 |
0.0767662958 |
- |
- |
dynamin, putative [Ricinus communis] |
| 9 |
Hb_001085_110 |
0.0768203535 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 10 |
Hb_004645_070 |
0.0776611738 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 11 |
Hb_005723_020 |
0.0778120198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004129_070 |
0.0785010258 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 13 |
Hb_000139_130 |
0.0795974403 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 14 |
Hb_000336_020 |
0.079848409 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 15 |
Hb_000066_150 |
0.0811949978 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_004109_370 |
0.0813986957 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_000479_240 |
0.0818631968 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008253_020 |
0.0829811217 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 19 |
Hb_031527_030 |
0.0831049748 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 20 |
Hb_010315_020 |
0.0838302615 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |