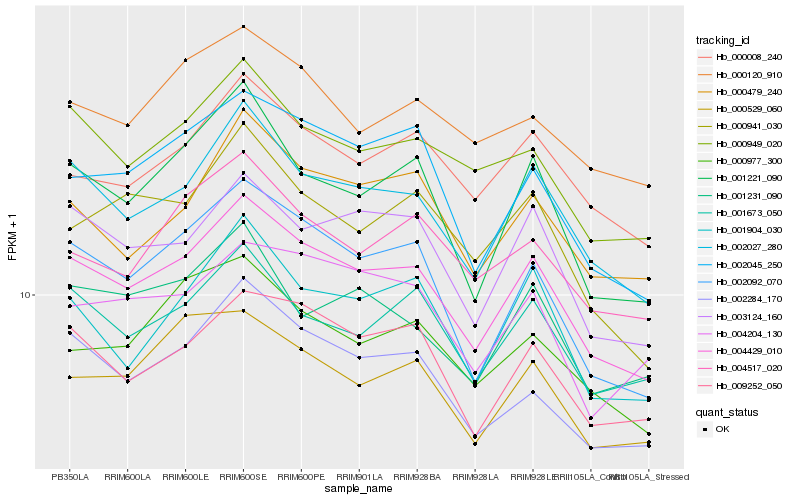
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004429_010 |
0.0 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 2 |
Hb_002027_280 |
0.0360789844 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 3 |
Hb_002092_070 |
0.0532140253 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 4 |
Hb_009252_050 |
0.0540776285 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_003124_160 |
0.0549270778 |
- |
- |
dynamin, putative [Ricinus communis] |
| 6 |
Hb_002284_170 |
0.0570035947 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 7 |
Hb_001221_090 |
0.0585040078 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001673_050 |
0.0595105215 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000949_020 |
0.0599691199 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 10 |
Hb_002045_250 |
0.0612600961 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000008_240 |
0.0620852274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001904_030 |
0.0623855757 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 13 |
Hb_000479_240 |
0.0627791933 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000529_060 |
0.0657118105 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000977_300 |
0.0669452439 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 16 |
Hb_004517_020 |
0.0684123406 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 17 |
Hb_000941_030 |
0.0687676548 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 18 |
Hb_001231_090 |
0.0688571618 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000120_910 |
0.0696806177 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 20 |
Hb_004204_130 |
0.0707603396 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |