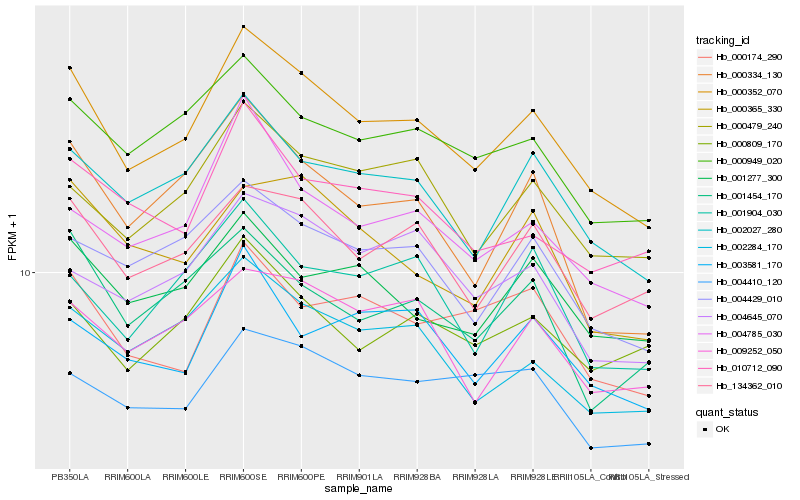
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_070 |
0.0 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 2 |
Hb_002027_280 |
0.0637223666 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 3 |
Hb_134362_010 |
0.0740291205 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 4 |
Hb_000949_020 |
0.0747504555 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 5 |
Hb_004429_010 |
0.0756756452 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 6 |
Hb_002284_170 |
0.0766915942 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 7 |
Hb_004785_030 |
0.0779901257 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 8 |
Hb_000479_240 |
0.0786675716 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_009252_050 |
0.0826505092 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001277_300 |
0.0836971404 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000334_130 |
0.0837444999 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 12 |
Hb_010712_090 |
0.084688884 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 13 |
Hb_001454_170 |
0.0873839416 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 14 |
Hb_000809_170 |
0.0880354968 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_003581_170 |
0.0880449856 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004645_070 |
0.0883380683 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 17 |
Hb_001904_030 |
0.088896421 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 18 |
Hb_000174_290 |
0.0889685849 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_004410_120 |
0.0891411984 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000365_330 |
0.0893423238 |
- |
- |
zinc finger protein, putative [Ricinus communis] |