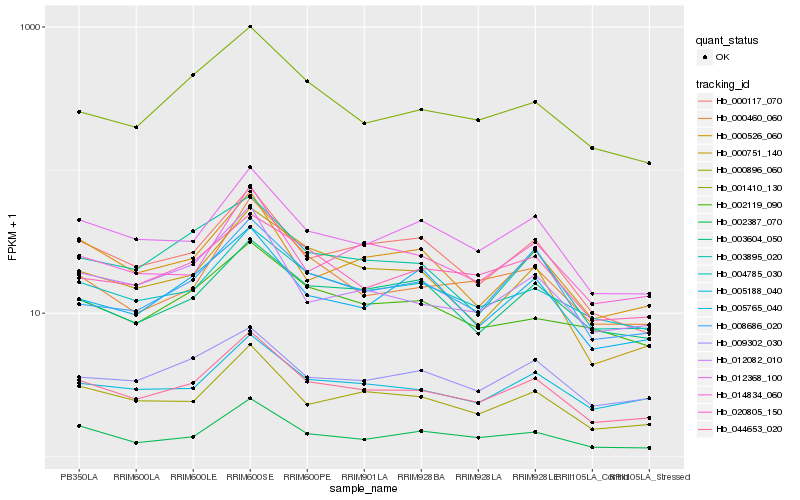
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044653_020 |
0.0 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 2 |
Hb_012368_100 |
0.0609226039 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 3 |
Hb_000896_060 |
0.0639027866 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_004785_030 |
0.0676904042 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 5 |
Hb_000117_070 |
0.0737308296 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 6 |
Hb_005188_040 |
0.0744109875 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 7 |
Hb_002387_070 |
0.077493663 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g77010, mitochondrial [Jatropha curcas] |
| 8 |
Hb_009302_030 |
0.0809658603 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 9 |
Hb_020805_150 |
0.0820905572 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000526_060 |
0.0825882498 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 11 |
Hb_003895_020 |
0.0832646122 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 12 |
Hb_014834_060 |
0.0840078493 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012082_010 |
0.0842115809 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001410_130 |
0.0849582926 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 15 |
Hb_000460_060 |
0.0862995571 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 16 |
Hb_002119_090 |
0.0873485686 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 17 |
Hb_003604_050 |
0.0876222781 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000751_140 |
0.0880127136 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 19 |
Hb_005765_040 |
0.0895067995 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_008686_020 |
0.0903523948 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |