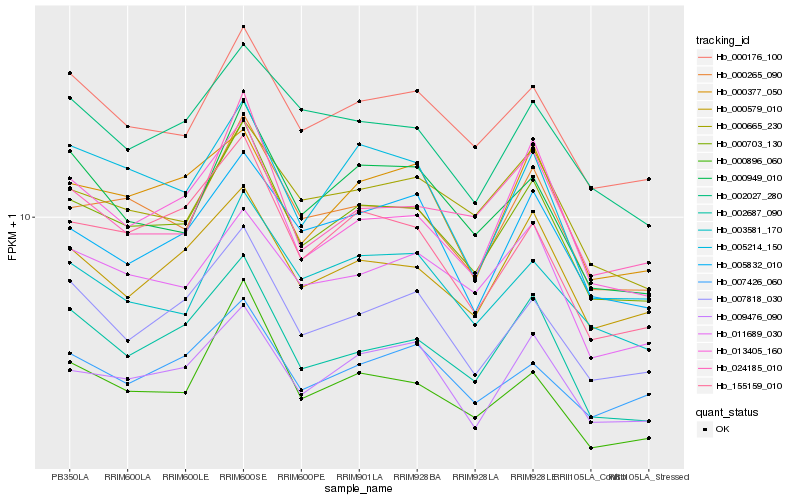
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_130 |
0.0 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 2 |
Hb_000265_090 |
0.0434623051 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 3 |
Hb_000176_100 |
0.0550186169 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 4 |
Hb_007818_030 |
0.0600237235 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 5 |
Hb_009476_090 |
0.0604622579 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_000579_010 |
0.0624887192 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 7 |
Hb_002687_090 |
0.0639246455 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_155159_010 |
0.0648237891 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 9 |
Hb_000665_230 |
0.0658769694 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 10 |
Hb_003581_170 |
0.0664875115 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000949_010 |
0.0671845046 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 12 |
Hb_013405_160 |
0.0676753889 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_011689_030 |
0.0689689692 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 14 |
Hb_005832_010 |
0.0694425286 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002027_280 |
0.0713239687 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 16 |
Hb_024185_010 |
0.0731060446 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007426_060 |
0.073183079 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_005214_150 |
0.0733215061 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 19 |
Hb_000896_060 |
0.0733247632 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000377_050 |
0.0736245288 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |