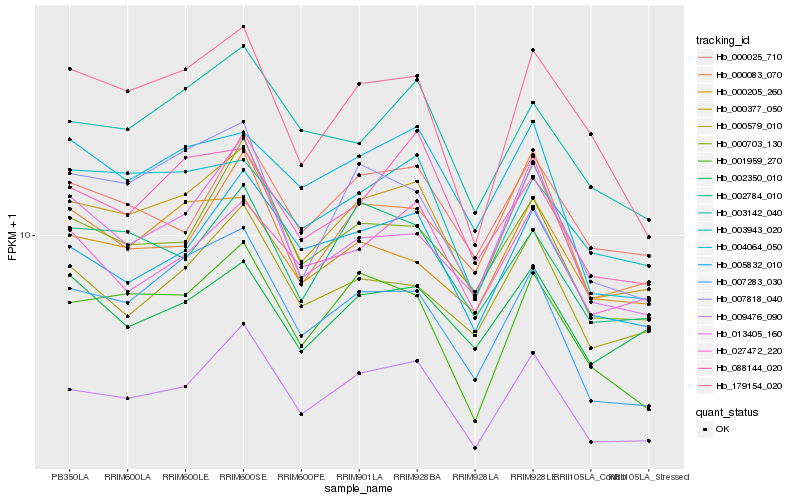
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000377_050 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 2 |
Hb_009476_090 |
0.0412697996 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 3 |
Hb_007283_030 |
0.0704824179 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 4 |
Hb_005832_010 |
0.0723771939 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001959_270 |
0.0728460594 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_007818_040 |
0.0731216454 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000703_130 |
0.0736245288 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 8 |
Hb_000579_010 |
0.0740766787 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 9 |
Hb_002784_010 |
0.0748427471 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 10 |
Hb_000025_710 |
0.0751145799 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 11 |
Hb_000083_070 |
0.0764537884 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003943_020 |
0.077220485 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_179154_020 |
0.0774453981 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_002350_010 |
0.0775141596 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 15 |
Hb_004064_050 |
0.0777864553 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 16 |
Hb_000205_260 |
0.077961873 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 17 |
Hb_027472_220 |
0.0780949066 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_003142_040 |
0.0782003424 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 19 |
Hb_088144_020 |
0.0784897589 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_013405_160 |
0.0788179449 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |