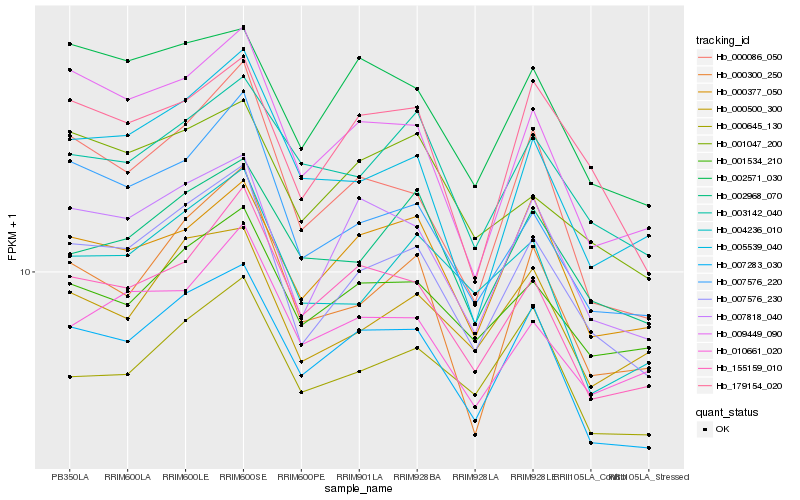
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_230 |
0.0 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 2 |
Hb_007818_040 |
0.0632947005 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007283_030 |
0.0678842525 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 4 |
Hb_007576_220 |
0.068694426 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 5 |
Hb_000086_050 |
0.0773648026 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 6 |
Hb_009449_090 |
0.0792283365 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000300_250 |
0.0841358817 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 8 |
Hb_179154_020 |
0.084762413 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_000500_300 |
0.0848882579 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 10 |
Hb_001534_210 |
0.0850470859 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 11 |
Hb_155159_010 |
0.0856103981 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 12 |
Hb_002571_030 |
0.0879025205 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 13 |
Hb_005539_040 |
0.0883095841 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 14 |
Hb_004236_010 |
0.0897408662 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 15 |
Hb_001047_200 |
0.0909901913 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 16 |
Hb_002968_070 |
0.0922342104 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 17 |
Hb_000377_050 |
0.0922388718 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 18 |
Hb_010661_020 |
0.0934461423 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000645_130 |
0.0946475058 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_003142_040 |
0.0963583836 |
- |
- |
Sf3a3 [Gossypium arboreum] |