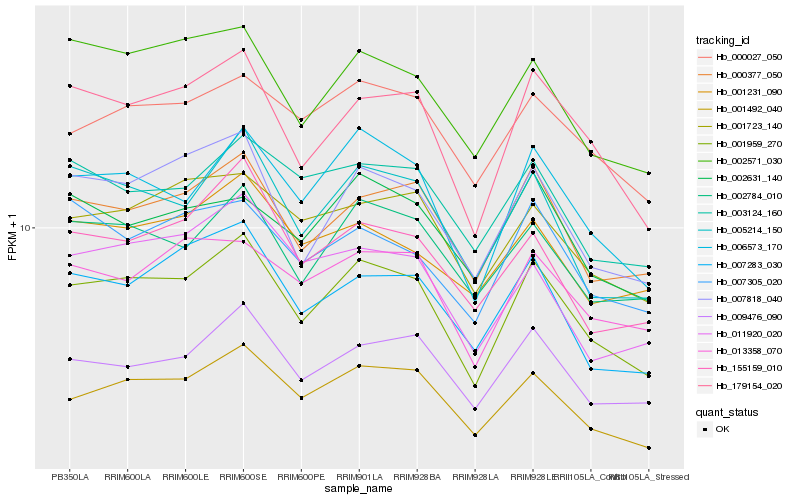
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_270 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_006573_170 |
0.0585825229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_179154_020 |
0.0590709484 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_001492_040 |
0.0628807393 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007818_040 |
0.0686983491 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005214_150 |
0.071106571 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 7 |
Hb_000377_050 |
0.0728460594 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 8 |
Hb_009476_090 |
0.0735710115 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_002784_010 |
0.079913471 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 10 |
Hb_002571_030 |
0.081555942 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 11 |
Hb_007283_030 |
0.0816922346 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 12 |
Hb_013358_070 |
0.0860835717 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_001723_140 |
0.0867783786 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_000027_050 |
0.0871318184 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 15 |
Hb_155159_010 |
0.0884737905 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 16 |
Hb_007305_020 |
0.0930525458 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0933368434 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_011920_020 |
0.0947899409 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 19 |
Hb_001231_090 |
0.0953236546 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002631_140 |
0.0964247651 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |