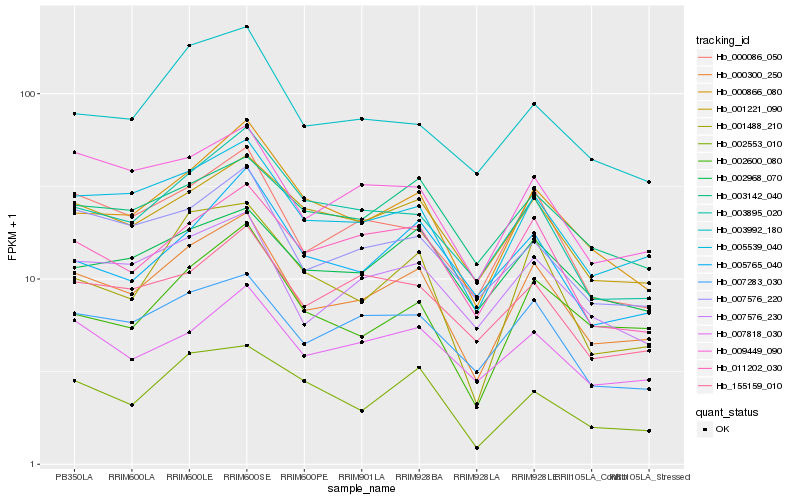
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_250 |
0.0 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 2 |
Hb_005539_040 |
0.0623194933 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000866_080 |
0.0795754858 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000086_050 |
0.0807538548 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 5 |
Hb_007576_230 |
0.0841358817 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 6 |
Hb_001221_090 |
0.0872458114 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009449_090 |
0.0892955163 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007283_030 |
0.0902004714 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 9 |
Hb_007576_220 |
0.0903080874 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 10 |
Hb_011202_030 |
0.0923487931 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 11 |
Hb_001488_210 |
0.096112494 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 12 |
Hb_003895_020 |
0.0967554788 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007818_030 |
0.0984387935 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 14 |
Hb_002600_080 |
0.0986487489 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_155159_010 |
0.0993961238 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 16 |
Hb_003992_180 |
0.1002458599 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002968_070 |
0.1029377506 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 18 |
Hb_005765_040 |
0.1043950799 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_002553_010 |
0.1044661636 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 20 |
Hb_003142_040 |
0.1050375602 |
- |
- |
Sf3a3 [Gossypium arboreum] |