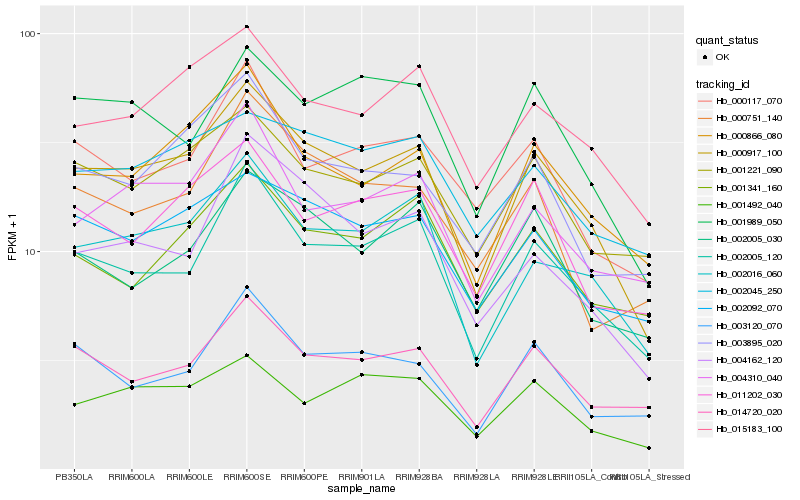
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_100 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 2 |
Hb_002005_120 |
0.0789460124 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 3 |
Hb_000866_080 |
0.0839457946 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002016_060 |
0.0842632598 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 5 |
Hb_015183_100 |
0.0920659003 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 6 |
Hb_002092_070 |
0.0971451954 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 7 |
Hb_001221_090 |
0.0999290183 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 8 |
Hb_014720_020 |
0.1002105538 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 9 |
Hb_004162_120 |
0.1038669538 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 10 |
Hb_003895_020 |
0.1047639651 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004310_040 |
0.1065622704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011202_030 |
0.1072003753 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 13 |
Hb_001989_050 |
0.1087596607 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 14 |
Hb_001492_040 |
0.1107324251 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000751_140 |
0.1127153305 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 16 |
Hb_002045_250 |
0.1127223316 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000117_070 |
0.112794293 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 18 |
Hb_002005_030 |
0.113743776 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_003120_070 |
0.1139383499 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_001341_160 |
0.1149990213 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |