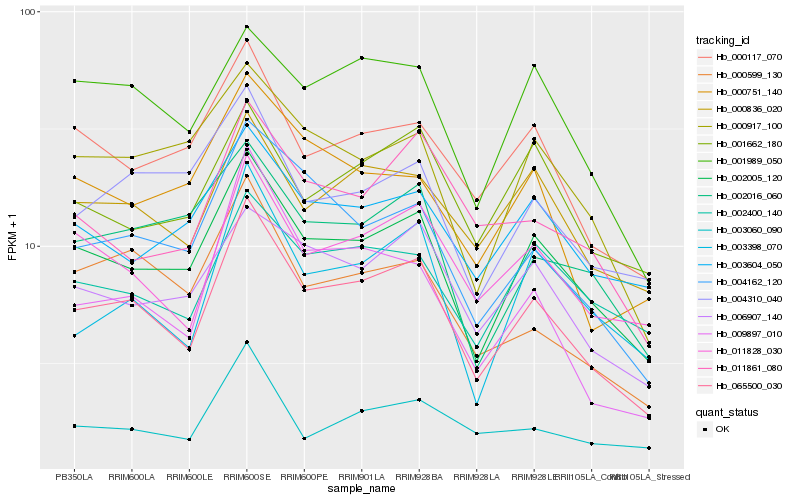
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065500_030 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 2 |
Hb_002005_120 |
0.0717536056 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 3 |
Hb_011828_030 |
0.0955008551 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 4 |
Hb_004162_120 |
0.097653866 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 5 |
Hb_000836_020 |
0.1044399351 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 6 |
Hb_000117_070 |
0.1096026323 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 7 |
Hb_000599_130 |
0.1106130477 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_006907_140 |
0.1109742413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009897_010 |
0.112606903 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_003398_070 |
0.1130058995 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 11 |
Hb_001989_050 |
0.1130858578 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 12 |
Hb_002016_060 |
0.1197804128 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_000917_100 |
0.119886409 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 14 |
Hb_004310_040 |
0.1200801573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002400_140 |
0.1205854176 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_011861_080 |
0.1211716734 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 17 |
Hb_003604_050 |
0.1247432584 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001662_180 |
0.1252039528 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 19 |
Hb_003060_090 |
0.1252186068 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 20 |
Hb_000751_140 |
0.1252431888 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |