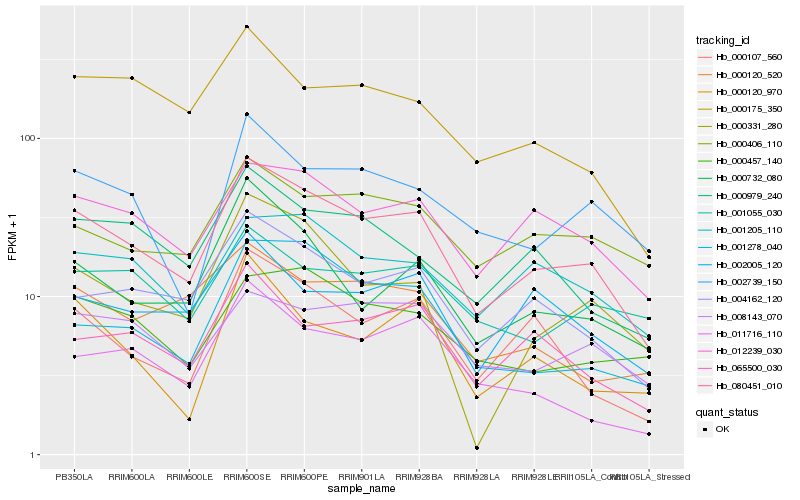
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080451_010 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 2 |
Hb_004162_120 |
0.1051192772 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 3 |
Hb_012239_030 |
0.1131456059 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000107_560 |
0.1244573011 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 5 |
Hb_065500_030 |
0.1297470469 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 6 |
Hb_000979_240 |
0.1301198202 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 7 |
Hb_001205_110 |
0.1304981239 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 8 |
Hb_000732_080 |
0.1323679072 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 9 |
Hb_002739_150 |
0.1343217115 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 10 |
Hb_000406_110 |
0.1354296303 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 11 |
Hb_000175_350 |
0.1359672294 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 12 |
Hb_002005_120 |
0.1371983177 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 13 |
Hb_001278_040 |
0.1378503499 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_011716_110 |
0.1389919064 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 15 |
Hb_000457_140 |
0.1404833963 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 16 |
Hb_008143_070 |
0.1409919163 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 17 |
Hb_000120_520 |
0.1429476221 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 18 |
Hb_001055_030 |
0.1453155962 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000331_280 |
0.1453179798 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 20 |
Hb_000120_970 |
0.1481829875 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |