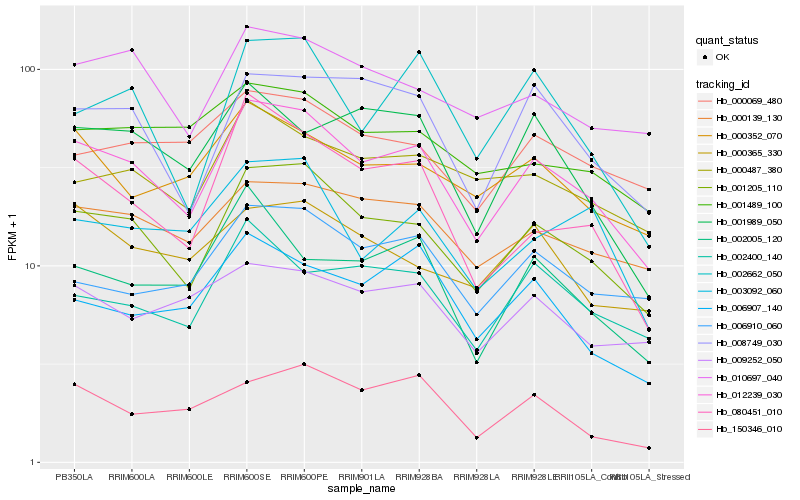
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012239_030 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001205_110 |
0.0457155113 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 3 |
Hb_000139_130 |
0.1025143241 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 4 |
Hb_008749_030 |
0.1064443081 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 5 |
Hb_010697_040 |
0.107649669 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000352_070 |
0.1083444578 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_080451_010 |
0.1131456059 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 8 |
Hb_002400_140 |
0.1157148531 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000487_380 |
0.1159893034 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006910_060 |
0.1175700319 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 11 |
Hb_000069_480 |
0.1185563705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001489_100 |
0.1187416108 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 13 |
Hb_006907_140 |
0.1193858744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009252_050 |
0.1198425256 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003092_060 |
0.1202719749 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_001989_050 |
0.12155073 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 17 |
Hb_002662_050 |
0.121560468 |
- |
- |
cysteine protease CP1 [Manihot esculenta] |
| 18 |
Hb_000365_330 |
0.1218113686 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_150346_010 |
0.1224902737 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_002005_120 |
0.1231973946 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |