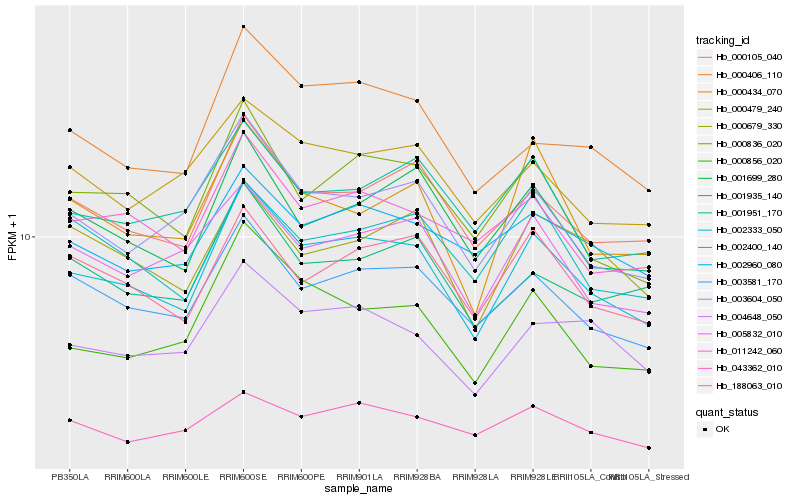
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_140 |
0.0 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 2 |
Hb_003581_170 |
0.0692131217 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000105_040 |
0.0739321205 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 4 |
Hb_000836_020 |
0.0746946194 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 5 |
Hb_003604_050 |
0.0784087745 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_004648_050 |
0.0809783307 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 7 |
Hb_188063_010 |
0.0815402515 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 8 |
Hb_000406_110 |
0.0816637499 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 9 |
Hb_001699_280 |
0.0824416562 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 10 |
Hb_011242_060 |
0.0830176764 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 11 |
Hb_000856_020 |
0.0843258847 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 12 |
Hb_005832_010 |
0.0855281251 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002960_080 |
0.0887564631 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 14 |
Hb_000434_070 |
0.0893996569 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 15 |
Hb_001935_140 |
0.0903826093 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 16 |
Hb_001951_170 |
0.090870961 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 17 |
Hb_000479_240 |
0.0915455774 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_043362_010 |
0.0923813191 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 19 |
Hb_002333_050 |
0.0925491609 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000679_330 |
0.0936289836 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |