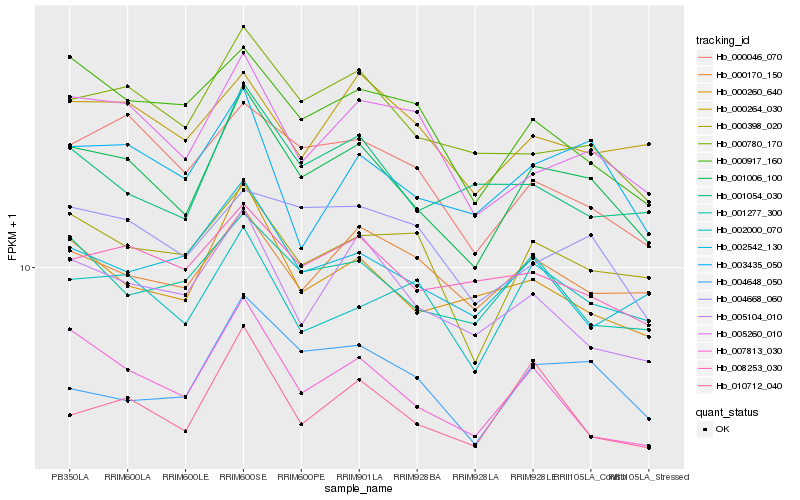
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_100 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000780_170 |
0.0882298471 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 3 |
Hb_000260_640 |
0.0897175921 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 4 |
Hb_005260_010 |
0.091381007 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000398_020 |
0.0935235664 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001277_300 |
0.0947279903 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 7 |
Hb_000046_070 |
0.0948366575 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 8 |
Hb_004648_050 |
0.095704438 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_007813_030 |
0.0957787695 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 10 |
Hb_008253_030 |
0.0985140513 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 11 |
Hb_000170_150 |
0.0998135839 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 12 |
Hb_005104_010 |
0.1001818093 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 13 |
Hb_004668_060 |
0.1016420496 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003435_050 |
0.1017460695 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1-like [Jatropha curcas] |
| 15 |
Hb_000917_160 |
0.1019293024 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001054_030 |
0.1029156907 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 17 |
Hb_002542_130 |
0.1034010928 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 18 |
Hb_000264_030 |
0.1035680161 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 19 |
Hb_010712_040 |
0.1036778137 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290 [Jatropha curcas] |
| 20 |
Hb_002000_070 |
0.1037130676 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |