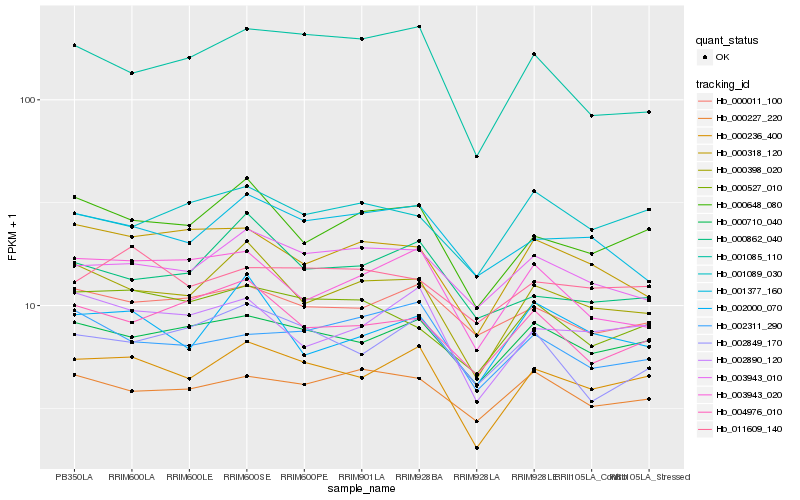
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_400 |
0.0 |
- |
- |
hypothetical protein JCGZ_01058 [Jatropha curcas] |
| 2 |
Hb_000710_040 |
0.0721434883 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 3 |
Hb_002890_120 |
0.0743888553 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 4 |
Hb_000648_080 |
0.0744334446 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 5 |
Hb_000398_020 |
0.0754567978 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000011_100 |
0.0921522105 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 7 |
Hb_003943_010 |
0.0939235281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000527_010 |
0.096221103 |
- |
- |
katanin P80 subunit, putative [Ricinus communis] |
| 9 |
Hb_001085_110 |
0.0972406483 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 10 |
Hb_004976_010 |
0.0978778481 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 11 |
Hb_001377_160 |
0.0983161983 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 12 |
Hb_002311_290 |
0.0987401437 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 13 |
Hb_003943_020 |
0.0992295111 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011609_140 |
0.100494537 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 15 |
Hb_002849_170 |
0.1006578601 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000227_220 |
0.1011004054 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 17 |
Hb_002000_070 |
0.1011662273 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 18 |
Hb_000862_040 |
0.1014670772 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_001089_030 |
0.1015912349 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 20 |
Hb_000318_120 |
0.1015945437 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |