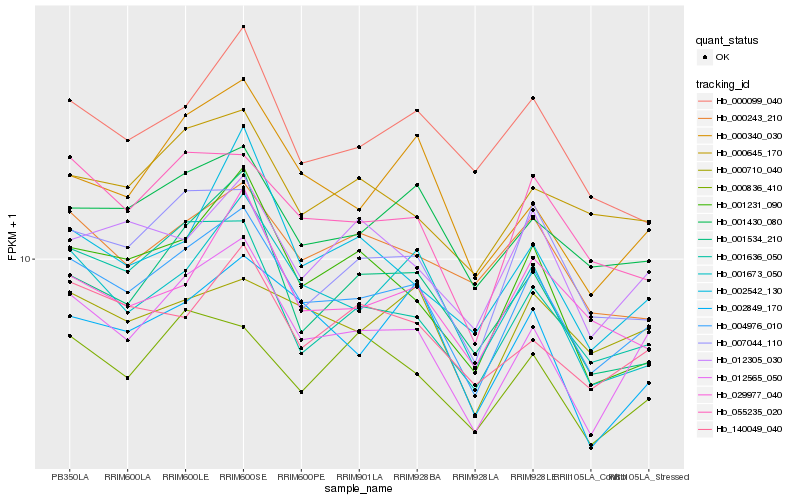
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004976_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 2 |
Hb_012565_050 |
0.0409038379 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 3 |
Hb_055235_020 |
0.0529993692 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001430_080 |
0.0548707188 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 5 |
Hb_007044_110 |
0.0564271874 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001673_050 |
0.0572202969 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001636_050 |
0.0618438733 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 8 |
Hb_001231_090 |
0.0624790327 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 9 |
Hb_140049_040 |
0.0635118215 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 10 |
Hb_000645_170 |
0.0637419216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_029977_040 |
0.0638889677 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 12 |
Hb_012305_030 |
0.0650900704 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 13 |
Hb_000836_410 |
0.0654407293 |
- |
- |
sec10, putative [Ricinus communis] |
| 14 |
Hb_002849_170 |
0.0659502544 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000340_030 |
0.0661880703 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 16 |
Hb_001534_210 |
0.0666878535 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000243_210 |
0.0668003364 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000099_040 |
0.0675253701 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000710_040 |
0.0678391108 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_002542_130 |
0.0689320466 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |