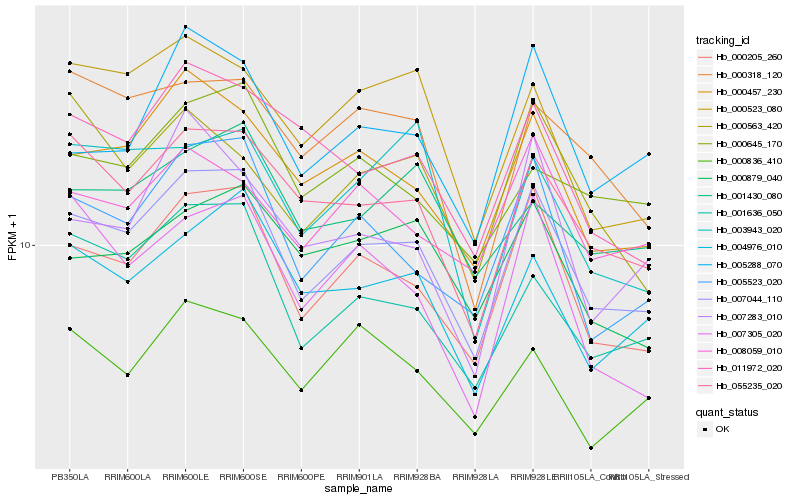
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_110 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000205_260 |
0.0328647404 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 3 |
Hb_000457_230 |
0.0489446377 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_055235_020 |
0.0505997632 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005523_020 |
0.0512165626 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 6 |
Hb_001636_050 |
0.0524224241 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 7 |
Hb_004976_010 |
0.0564271874 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 8 |
Hb_005288_070 |
0.0628131284 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_011972_020 |
0.066404632 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_008059_010 |
0.0665264027 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000523_080 |
0.0675112801 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000879_040 |
0.0682281233 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_000836_410 |
0.0684536284 |
- |
- |
sec10, putative [Ricinus communis] |
| 14 |
Hb_000645_170 |
0.0690365765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000318_120 |
0.0690612357 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001430_080 |
0.0696371561 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 17 |
Hb_007283_010 |
0.071963058 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 18 |
Hb_007305_020 |
0.0720017217 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 19 |
Hb_000563_420 |
0.0737196491 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 20 |
Hb_003943_020 |
0.0740335647 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |