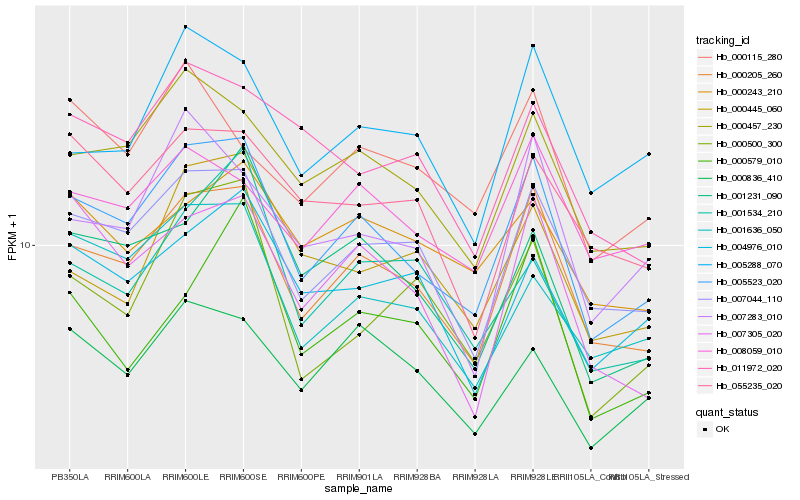
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005523_020 |
0.0 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 2 |
Hb_000205_260 |
0.0327607593 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 3 |
Hb_007044_110 |
0.0512165626 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_000457_230 |
0.0539343302 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001636_050 |
0.0657892845 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 6 |
Hb_001231_090 |
0.0665737192 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007305_020 |
0.0720073341 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 8 |
Hb_000836_410 |
0.0740325776 |
- |
- |
sec10, putative [Ricinus communis] |
| 9 |
Hb_004976_010 |
0.0745462894 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 10 |
Hb_007283_010 |
0.0746528195 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 11 |
Hb_005288_070 |
0.0749037555 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000500_300 |
0.0759987151 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 13 |
Hb_008059_010 |
0.0763475869 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001534_210 |
0.0775982012 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000115_280 |
0.0781278476 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_011972_020 |
0.0789758439 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_000579_010 |
0.0789884005 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 18 |
Hb_055235_020 |
0.0792728822 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000243_210 |
0.0795140359 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000445_060 |
0.0797316915 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |