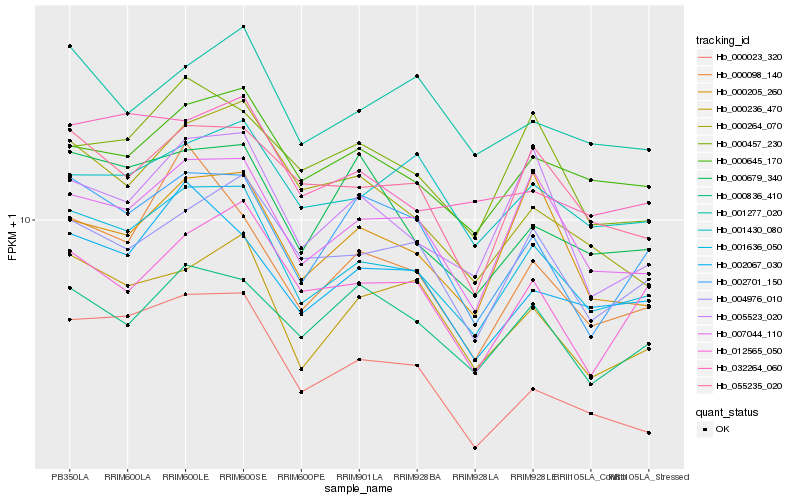
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001636_050 |
0.0 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 2 |
Hb_007044_110 |
0.0524224241 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002067_030 |
0.0599148391 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004976_010 |
0.0618438733 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 5 |
Hb_000645_170 |
0.062178825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000236_470 |
0.0623765514 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 7 |
Hb_055235_020 |
0.0631573748 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005523_020 |
0.0657892845 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 9 |
Hb_000023_320 |
0.0668046878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002701_150 |
0.0681410753 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000457_230 |
0.0682088123 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000679_340 |
0.0688518122 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 13 |
Hb_032264_060 |
0.0689102093 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 14 |
Hb_000098_140 |
0.0704607509 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 15 |
Hb_000836_410 |
0.07094052 |
- |
- |
sec10, putative [Ricinus communis] |
| 16 |
Hb_000205_260 |
0.071669126 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 17 |
Hb_000264_070 |
0.0722745466 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 18 |
Hb_001430_080 |
0.0737808064 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 19 |
Hb_012565_050 |
0.0741049436 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 20 |
Hb_001277_020 |
0.0749410033 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |