| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
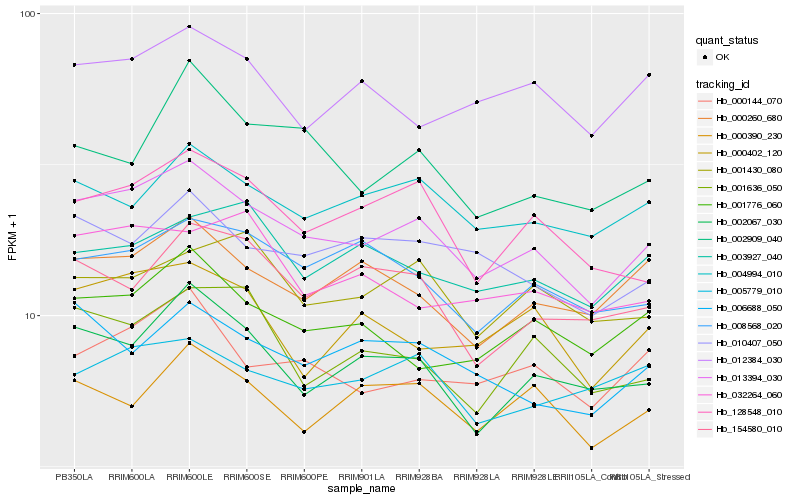
Hb_013394_030 |
0.0 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 2 |
Hb_002067_030 |
0.06929936 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_128548_010 |
0.0696869305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000144_070 |
0.0705185372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004994_010 |
0.0726187766 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_010407_050 |
0.0728342062 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 7 |
Hb_008568_020 |
0.0742073294 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 8 |
Hb_000260_680 |
0.0742737843 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 9 |
Hb_006688_050 |
0.0764615558 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001776_060 |
0.078301688 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 11 |
Hb_000390_230 |
0.0794986188 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_000402_120 |
0.0796430211 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 13 |
Hb_154580_010 |
0.0806122409 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 14 |
Hb_002909_040 |
0.0816069734 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 15 |
Hb_001636_050 |
0.0816491549 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 16 |
Hb_005779_010 |
0.0817573293 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_001430_080 |
0.0823873408 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 18 |
Hb_003927_040 |
0.0831939384 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 19 |
Hb_012384_030 |
0.0838780763 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 20 |
Hb_032264_060 |
0.0847721215 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |