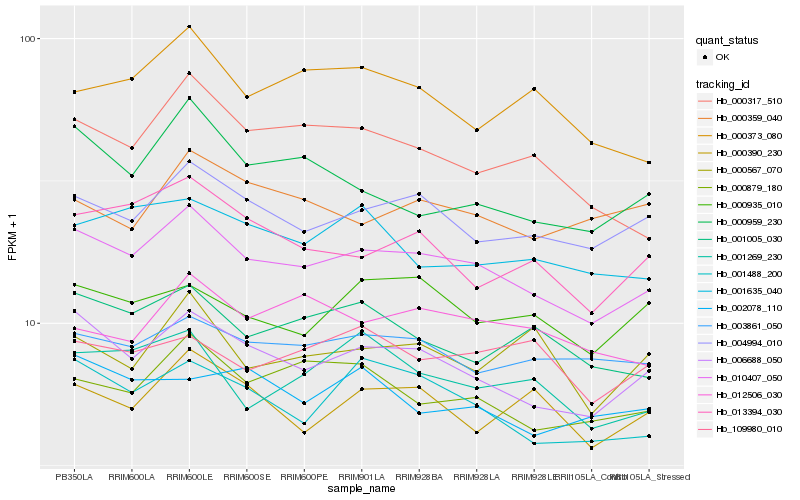
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_050 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 2 |
Hb_006688_050 |
0.0439095361 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004994_010 |
0.0578534011 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_001488_200 |
0.0633214955 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000879_180 |
0.0665968054 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_003861_050 |
0.0703200027 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 7 |
Hb_001005_030 |
0.0712339061 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_013394_030 |
0.0728342062 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 9 |
Hb_000317_510 |
0.0749040381 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 10 |
Hb_001269_230 |
0.0753167008 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 11 |
Hb_001635_040 |
0.0755160815 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 12 |
Hb_000390_230 |
0.0772082543 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 13 |
Hb_000935_010 |
0.0773622138 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 14 |
Hb_000959_230 |
0.077952452 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 15 |
Hb_012506_030 |
0.0800099185 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 16 |
Hb_000373_080 |
0.0815320684 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 17 |
Hb_000359_040 |
0.0831880217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_109980_010 |
0.0834903943 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002078_110 |
0.0835756048 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 20 |
Hb_000567_070 |
0.0839111671 |
- |
- |
autophagy protein, putative [Ricinus communis] |