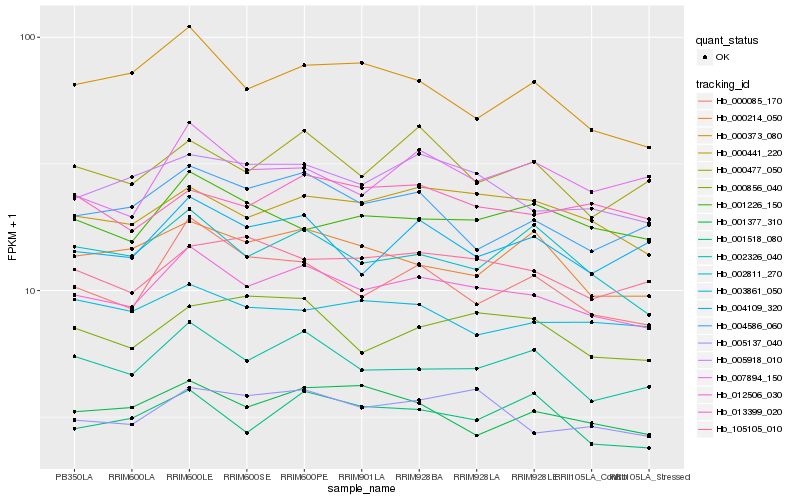
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012506_030 |
0.0 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 2 |
Hb_002326_040 |
0.0507576627 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 3 |
Hb_000441_220 |
0.0520882105 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_005137_040 |
0.0553061247 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 5 |
Hb_005918_010 |
0.0564342899 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 6 |
Hb_000085_170 |
0.0604924675 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002811_270 |
0.0635781219 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 8 |
Hb_105105_010 |
0.0647293972 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 9 |
Hb_001226_150 |
0.0659364778 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_004109_320 |
0.066325904 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 11 |
Hb_001377_310 |
0.0667610241 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 12 |
Hb_004586_060 |
0.0672767949 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 13 |
Hb_000477_050 |
0.0673572938 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 14 |
Hb_001518_080 |
0.0679864145 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 15 |
Hb_013399_020 |
0.0684258539 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 16 |
Hb_000214_050 |
0.0686911449 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000856_040 |
0.069071239 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 18 |
Hb_000373_080 |
0.0702977492 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 19 |
Hb_003861_050 |
0.0703832996 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 20 |
Hb_007894_150 |
0.0708701018 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |