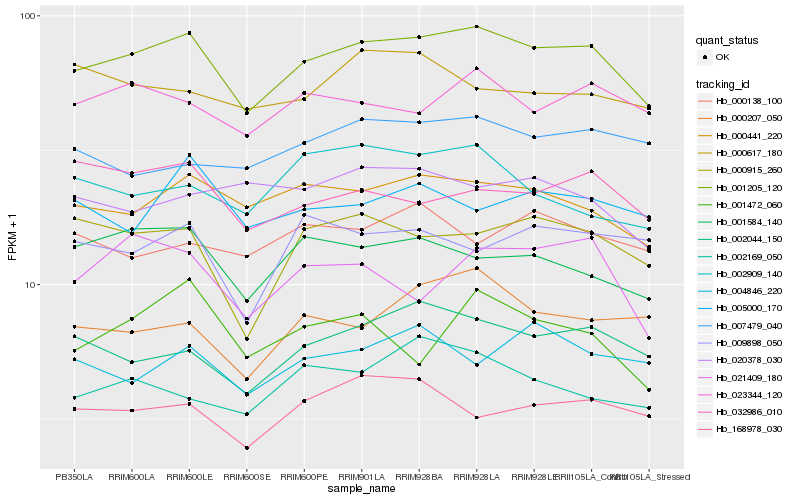
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_120 |
0.0 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 2 |
Hb_000441_220 |
0.0636696014 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002044_150 |
0.0712025816 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 4 |
Hb_001584_140 |
0.0730812112 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 5 |
Hb_023344_120 |
0.0739812 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000915_260 |
0.0772098562 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 7 |
Hb_168978_030 |
0.0799514154 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 8 |
Hb_032986_010 |
0.0815811142 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 9 |
Hb_005000_170 |
0.083748869 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 10 |
Hb_000207_050 |
0.0837739155 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021409_180 |
0.0842421627 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 12 |
Hb_002169_050 |
0.0856572945 |
- |
- |
PREDICTED: inositol 1,3,4-trisphosphate 5/6-kinase 4 [Jatropha curcas] |
| 13 |
Hb_007479_040 |
0.0859245459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002909_140 |
0.0867728878 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 2 [Jatropha curcas] |
| 15 |
Hb_000617_180 |
0.0873281088 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 16 |
Hb_004846_220 |
0.0874157417 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009898_050 |
0.0875770526 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 18 |
Hb_020378_030 |
0.0880617763 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 19 |
Hb_000138_100 |
0.0881085504 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 20 |
Hb_001472_060 |
0.088339211 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |