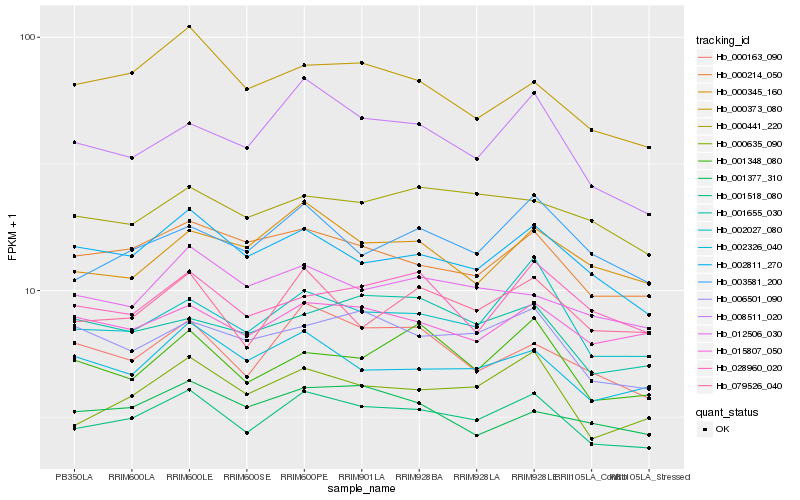
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001518_080 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 2 |
Hb_000635_090 |
0.0564343024 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 3 |
Hb_000214_050 |
0.0587601685 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_079526_040 |
0.0587899592 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 5 |
Hb_002027_080 |
0.059077813 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 6 |
Hb_002811_270 |
0.0593555899 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 7 |
Hb_015807_050 |
0.0631834115 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 8 |
Hb_002326_040 |
0.0647980236 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 9 |
Hb_000163_090 |
0.0653135498 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 10 |
Hb_003581_200 |
0.0655260886 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 11 |
Hb_006501_090 |
0.0662131423 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 12 |
Hb_008511_020 |
0.0666486925 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 13 |
Hb_000441_220 |
0.0675464312 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_012506_030 |
0.0679864145 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 15 |
Hb_000345_160 |
0.0701740846 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_001348_080 |
0.0703410251 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_001655_030 |
0.0712308044 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000373_080 |
0.0717243397 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 19 |
Hb_028960_020 |
0.0718963863 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001377_310 |
0.0739533826 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |