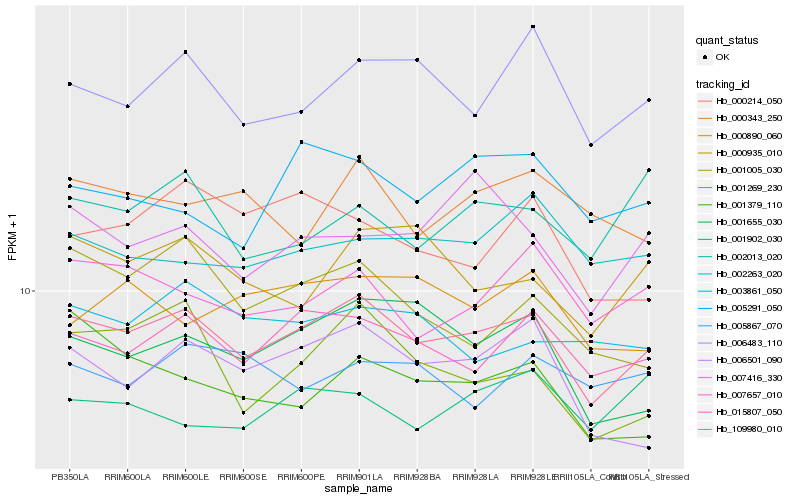
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_109980_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 2 |
Hb_015807_050 |
0.0492952172 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 3 |
Hb_006483_110 |
0.0618525502 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 4 |
Hb_006501_090 |
0.0647448631 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 5 |
Hb_001655_030 |
0.0648724338 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007657_010 |
0.0658522795 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000935_010 |
0.0680882243 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 8 |
Hb_002263_020 |
0.0701892013 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 9 |
Hb_001269_230 |
0.07027826 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 10 |
Hb_003861_050 |
0.0718104391 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 11 |
Hb_005291_050 |
0.0724383107 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_007416_330 |
0.0726889837 |
- |
- |
PREDICTED: elongation factor 1-alpha isoform X1 [Jatropha curcas] |
| 13 |
Hb_002013_020 |
0.0733777878 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 14 |
Hb_000343_250 |
0.0744660175 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 15 |
Hb_001379_110 |
0.0748817762 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 16 |
Hb_001005_030 |
0.0750121645 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000214_050 |
0.0753613715 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001902_030 |
0.0754471346 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005867_070 |
0.0756027441 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000890_060 |
0.0760639027 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |