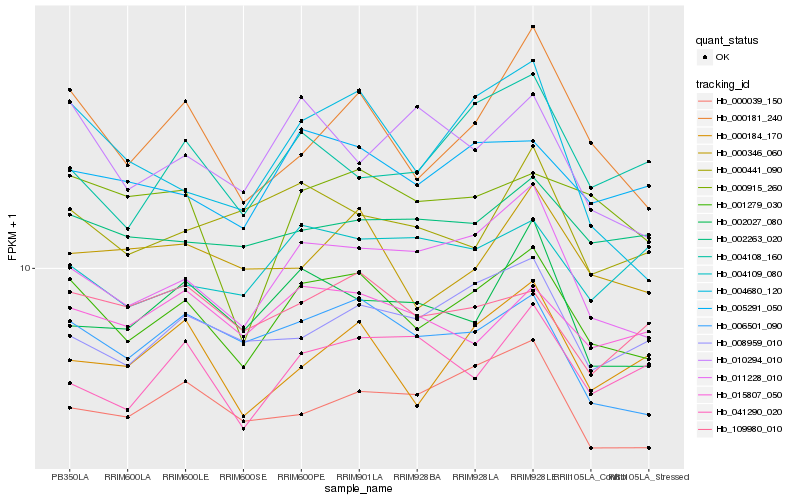
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 2 |
Hb_011228_010 |
0.0571375029 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_005291_050 |
0.0655996195 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_006501_090 |
0.0663180367 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 5 |
Hb_000181_240 |
0.0689271865 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 6 |
Hb_000039_150 |
0.073662827 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 7 |
Hb_015807_050 |
0.0768957858 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 8 |
Hb_008959_010 |
0.0772941865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_109980_010 |
0.0776102906 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002263_020 |
0.0779437261 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 11 |
Hb_000184_170 |
0.0780841312 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 12 |
Hb_000346_060 |
0.0784642146 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 13 |
Hb_002027_080 |
0.0808939256 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 14 |
Hb_004680_120 |
0.082096299 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 15 |
Hb_000915_260 |
0.082199923 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000441_090 |
0.0834349424 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 17 |
Hb_010294_010 |
0.0852188987 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 18 |
Hb_004108_160 |
0.0853407742 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 19 |
Hb_041290_020 |
0.0859212887 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 20 |
Hb_004109_080 |
0.0864086435 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |