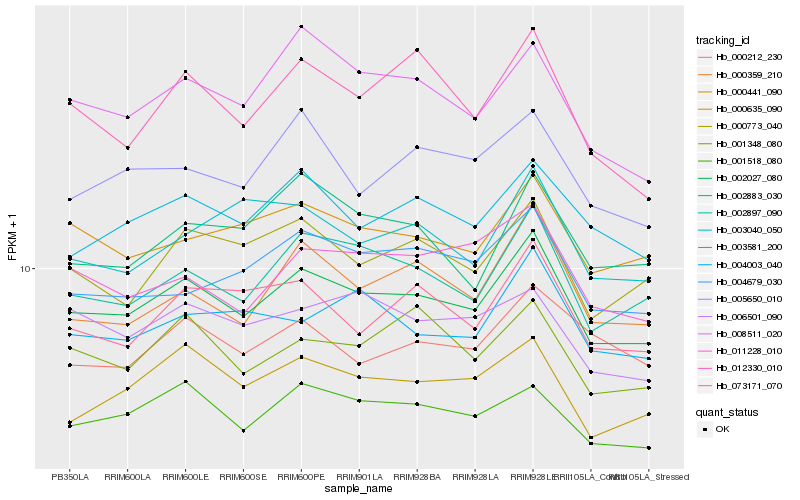
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_080 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 2 |
Hb_002897_090 |
0.0509366312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001518_080 |
0.059077813 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 4 |
Hb_003581_200 |
0.0604575438 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 5 |
Hb_000441_090 |
0.061709268 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 6 |
Hb_000773_040 |
0.0625340464 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000635_090 |
0.0626018091 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 8 |
Hb_000359_210 |
0.0630311484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_073171_070 |
0.0658704552 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 10 |
Hb_008511_020 |
0.0666061494 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 11 |
Hb_004679_030 |
0.0670007988 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 12 |
Hb_004003_040 |
0.068492997 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_011228_010 |
0.0687243158 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001348_080 |
0.0691679149 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 15 |
Hb_005650_010 |
0.0693994419 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_012330_010 |
0.072248962 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002883_030 |
0.0735510925 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 18 |
Hb_000212_230 |
0.0739446553 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 19 |
Hb_006501_090 |
0.0742691634 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 20 |
Hb_003040_050 |
0.0749231863 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |