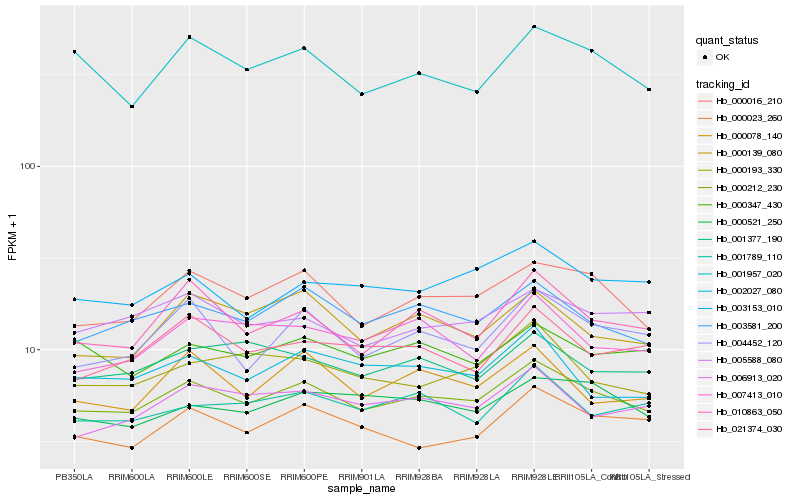
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_230 |
0.0 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 2 |
Hb_003581_200 |
0.0532836581 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 3 |
Hb_000016_210 |
0.0552092905 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 4 |
Hb_005588_080 |
0.0692577015 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 5 |
Hb_006913_020 |
0.0692652403 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010863_050 |
0.0696166655 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 7 |
Hb_000521_250 |
0.070765783 |
- |
- |
PREDICTED: protein N-terminal asparagine amidohydrolase isoform X2 [Jatropha curcas] |
| 8 |
Hb_001789_110 |
0.0715668155 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000023_260 |
0.0719112218 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001377_190 |
0.0720115185 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000347_430 |
0.0723217811 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 12 |
Hb_003153_010 |
0.0724495644 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 13 |
Hb_001957_020 |
0.0726874994 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 14 |
Hb_002027_080 |
0.0739446553 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 15 |
Hb_000139_080 |
0.0741730567 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004452_120 |
0.0744673038 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 17 |
Hb_021374_030 |
0.0745916161 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 18 |
Hb_007413_010 |
0.0750508194 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 19 |
Hb_000078_140 |
0.0751715942 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_000193_330 |
0.0760835591 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |