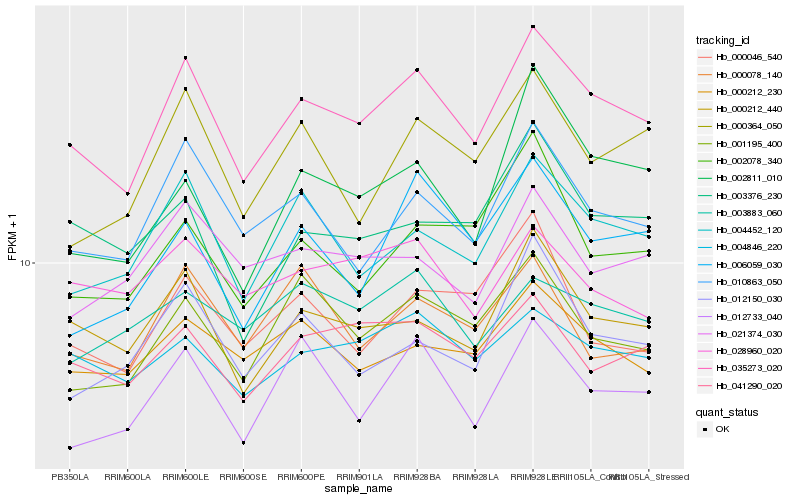
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035273_020 |
0.0 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000212_440 |
0.0660885843 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_004452_120 |
0.0706220839 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 4 |
Hb_010863_050 |
0.0707042277 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 5 |
Hb_001195_400 |
0.0759741976 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 6 |
Hb_004846_220 |
0.0791726724 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_041290_020 |
0.0798758215 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_012733_040 |
0.080327309 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 9 |
Hb_000212_230 |
0.0822256584 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 10 |
Hb_000364_050 |
0.0852585553 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_012150_030 |
0.0857332779 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002078_340 |
0.0881418814 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006059_030 |
0.0896149703 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_002811_010 |
0.090049858 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 15 |
Hb_021374_030 |
0.0910816382 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 16 |
Hb_000078_140 |
0.0913371454 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_003376_230 |
0.0915002034 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 18 |
Hb_000046_540 |
0.0915926993 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 19 |
Hb_028960_020 |
0.0926156459 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003883_060 |
0.0937493015 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |