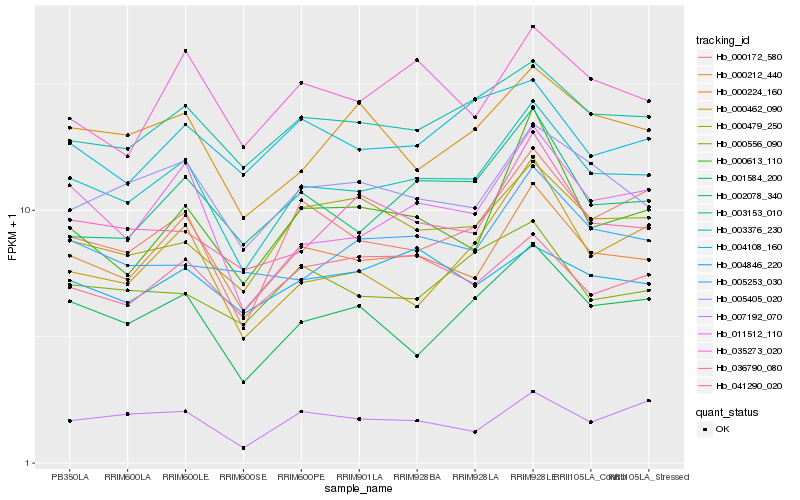
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_230 |
0.0 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 2 |
Hb_000212_440 |
0.0566183134 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_011512_110 |
0.0664428326 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_003153_010 |
0.0700028831 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 5 |
Hb_000172_580 |
0.0727155246 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001584_200 |
0.0747029567 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 7 |
Hb_005405_020 |
0.0778968416 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_002078_340 |
0.082639262 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000224_160 |
0.0852858625 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 10 |
Hb_000479_250 |
0.0856305038 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 11 |
Hb_036790_080 |
0.0872548178 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 12 |
Hb_035273_020 |
0.0915002034 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005253_030 |
0.0918543735 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000556_090 |
0.0942084694 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 15 |
Hb_041290_020 |
0.0947849977 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000613_110 |
0.0953003263 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 17 |
Hb_004108_160 |
0.0962804705 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 18 |
Hb_000462_090 |
0.0979409826 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_007192_070 |
0.098472509 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_004846_220 |
0.0985709593 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |