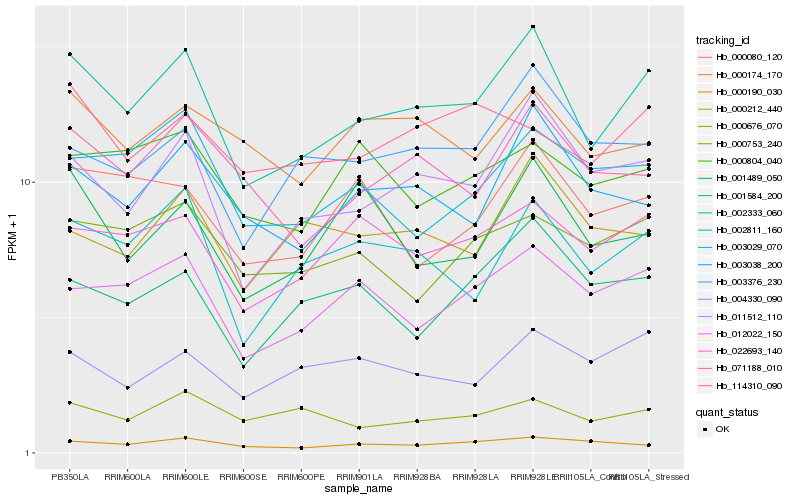
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002333_060 |
0.0 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_011512_110 |
0.0818337794 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002811_160 |
0.0940285938 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_022693_140 |
0.0971690335 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012022_150 |
0.0979665308 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 6 |
Hb_071188_010 |
0.1032424454 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 7 |
Hb_003038_200 |
0.1047348847 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 8 |
Hb_003376_230 |
0.1085782628 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 9 |
Hb_003029_070 |
0.114256956 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000174_170 |
0.115007614 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000753_240 |
0.1152302976 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 12 |
Hb_001584_200 |
0.1160910451 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 13 |
Hb_000190_030 |
0.1162902923 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 14 |
Hb_004330_090 |
0.1164052494 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000080_120 |
0.1174964155 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 16 |
Hb_000676_070 |
0.1181775442 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 17 |
Hb_000212_440 |
0.1187874092 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001489_050 |
0.1202184486 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_114310_090 |
0.1202813388 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase [Jatropha curcas] |
| 20 |
Hb_000804_040 |
0.1207288556 |
- |
- |
protein with unknown function [Ricinus communis] |