| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
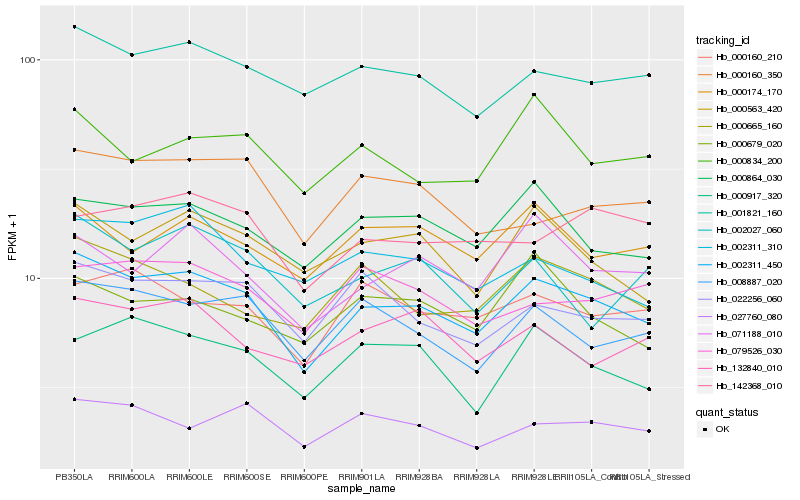
Hb_002311_450 |
0.0 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 2 |
Hb_001821_160 |
0.084241547 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 3 |
Hb_000864_030 |
0.0868404343 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008887_020 |
0.0890039737 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 5 |
Hb_000563_420 |
0.0906811153 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 6 |
Hb_022256_060 |
0.0927570744 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 7 |
Hb_071188_010 |
0.0933788627 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 8 |
Hb_000665_160 |
0.0958104947 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000160_210 |
0.0971647329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000917_320 |
0.097337076 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000174_170 |
0.0978222606 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_027760_080 |
0.0979105937 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_002311_310 |
0.0997142582 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 14 |
Hb_132840_010 |
0.100366244 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000160_350 |
0.1025847694 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 16 |
Hb_079526_030 |
0.103572893 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 17 |
Hb_000834_200 |
0.1039021461 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 18 |
Hb_002027_060 |
0.1045462697 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 19 |
Hb_142368_010 |
0.1048499941 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000679_020 |
0.1066893736 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |