| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
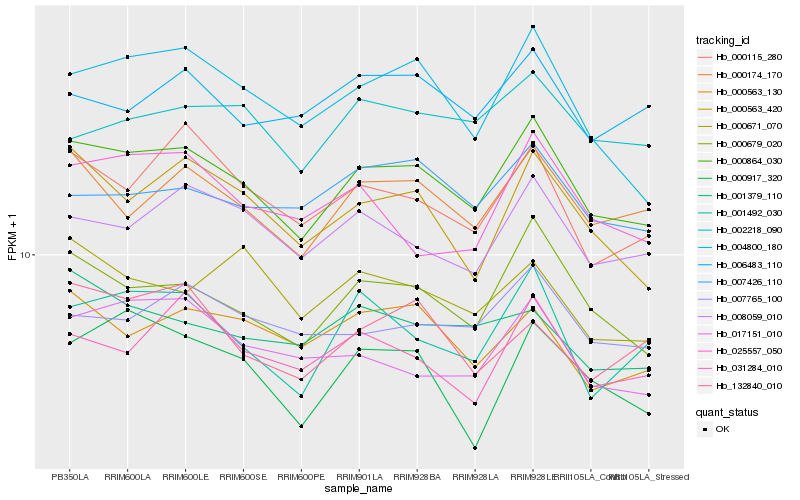
Hb_000864_030 |
0.0 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000679_020 |
0.0498441104 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 3 |
Hb_000174_170 |
0.0590085886 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004800_180 |
0.0636542428 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 5 |
Hb_008059_010 |
0.0673767773 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000563_420 |
0.0674773306 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 7 |
Hb_000115_280 |
0.0706115346 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 8 |
Hb_001492_030 |
0.0708626904 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007765_100 |
0.0728265793 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 10 |
Hb_000563_130 |
0.0755696419 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_031284_010 |
0.0758077501 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 12 |
Hb_017151_010 |
0.0776825705 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 13 |
Hb_007426_110 |
0.0780987815 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_002218_090 |
0.0800378603 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 15 |
Hb_006483_110 |
0.0802609204 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 16 |
Hb_025557_050 |
0.0806403757 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 17 |
Hb_001379_110 |
0.0809398824 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 18 |
Hb_000917_320 |
0.0822485098 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000671_070 |
0.0843176352 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 20 |
Hb_132840_010 |
0.0843698011 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |