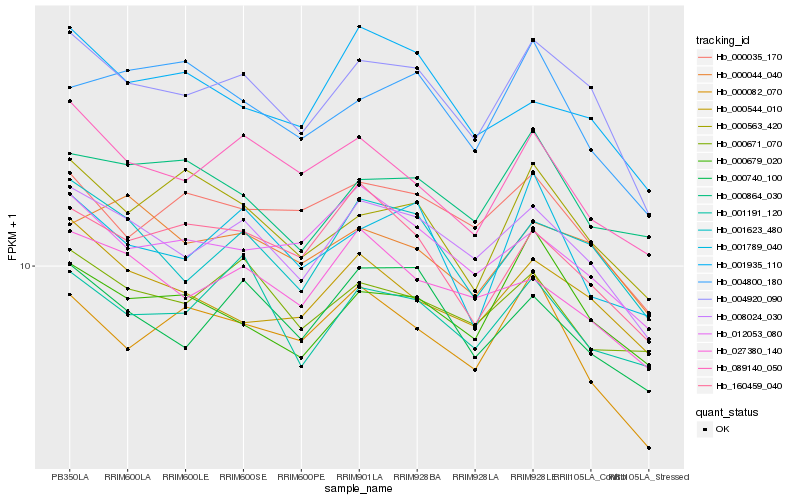
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004920_090 |
0.0 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_008024_030 |
0.0470651939 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 3 |
Hb_001623_480 |
0.0611125186 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 4 |
Hb_000679_020 |
0.0762982171 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 5 |
Hb_000563_420 |
0.0769731079 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 6 |
Hb_000035_170 |
0.077647386 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 7 |
Hb_000544_010 |
0.0789450281 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_089140_050 |
0.0796611272 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 9 |
Hb_000740_100 |
0.0813728547 |
- |
- |
calpain, putative [Ricinus communis] |
| 10 |
Hb_001191_120 |
0.0839466627 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000044_040 |
0.0842747714 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 12 |
Hb_012053_080 |
0.0843024146 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 13 |
Hb_000671_070 |
0.0852052149 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 14 |
Hb_001789_040 |
0.0853104065 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 15 |
Hb_027380_140 |
0.0865073506 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 16 |
Hb_160459_040 |
0.0874419563 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 17 |
Hb_000082_070 |
0.0874988388 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 18 |
Hb_000864_030 |
0.0893832992 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004800_180 |
0.0921087166 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 20 |
Hb_001935_110 |
0.0923183981 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |