| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
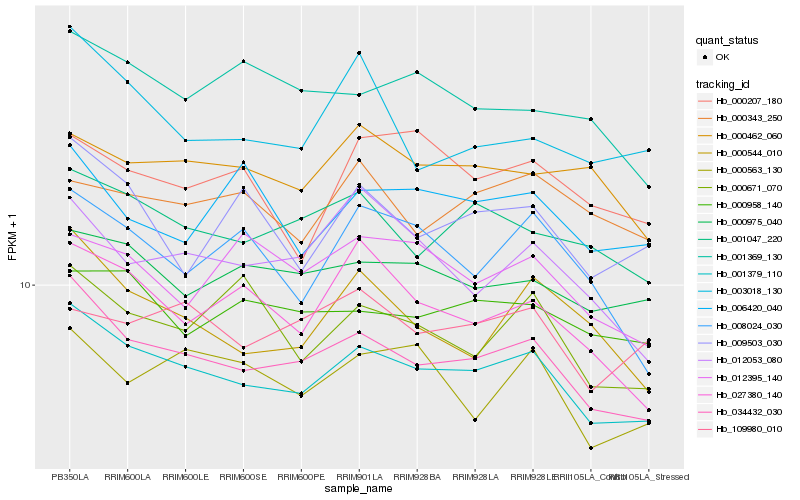
Hb_001379_110 |
0.0 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 2 |
Hb_034432_030 |
0.0270969823 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001047_220 |
0.0598990875 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 4 |
Hb_000975_040 |
0.0622803531 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 5 |
Hb_000958_140 |
0.067211817 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 6 |
Hb_000544_010 |
0.0677965092 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_027380_140 |
0.0702739363 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000671_070 |
0.0710051064 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 9 |
Hb_000462_060 |
0.0714853258 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000563_130 |
0.0722022192 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_009503_030 |
0.0727687994 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 12 |
Hb_001369_130 |
0.0742803529 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_109980_010 |
0.0748817762 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003018_130 |
0.0762053447 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 15 |
Hb_012053_080 |
0.0763245753 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 16 |
Hb_000207_180 |
0.0767013084 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 17 |
Hb_008024_030 |
0.0768912665 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 18 |
Hb_006420_040 |
0.0774723653 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000343_250 |
0.0775426909 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 20 |
Hb_012395_140 |
0.0778639606 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |