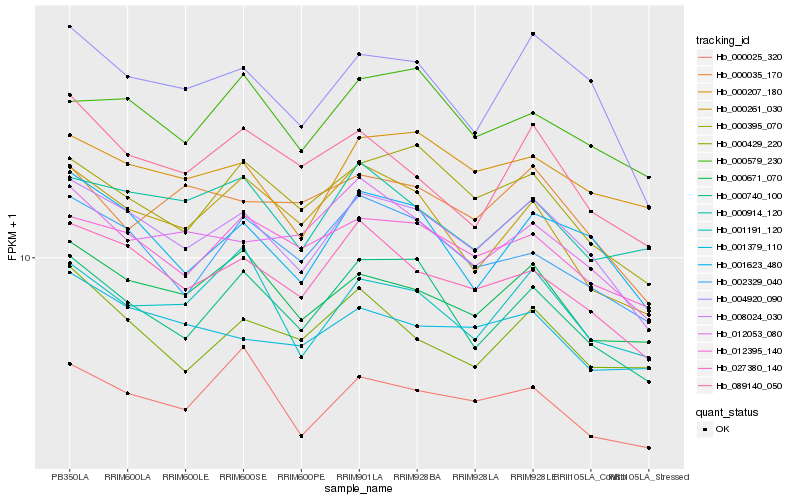
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008024_030 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 2 |
Hb_004920_090 |
0.0470651939 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_027380_140 |
0.0495681712 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001623_480 |
0.0529640087 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 5 |
Hb_000740_100 |
0.0593923577 |
- |
- |
calpain, putative [Ricinus communis] |
| 6 |
Hb_000671_070 |
0.0642954061 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 7 |
Hb_012395_140 |
0.0644413806 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 8 |
Hb_000395_070 |
0.0648588025 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002329_040 |
0.0651486152 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 10 |
Hb_012053_080 |
0.0707179731 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 11 |
Hb_000207_180 |
0.0720391207 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 12 |
Hb_000261_030 |
0.0723553657 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 13 |
Hb_000025_320 |
0.0730024961 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 14 |
Hb_001191_120 |
0.0736523666 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000579_230 |
0.0743564632 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 16 |
Hb_089140_050 |
0.075328748 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 17 |
Hb_001379_110 |
0.0768912665 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 18 |
Hb_000035_170 |
0.0778656341 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 19 |
Hb_000914_120 |
0.0778871848 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000429_220 |
0.0791407093 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |