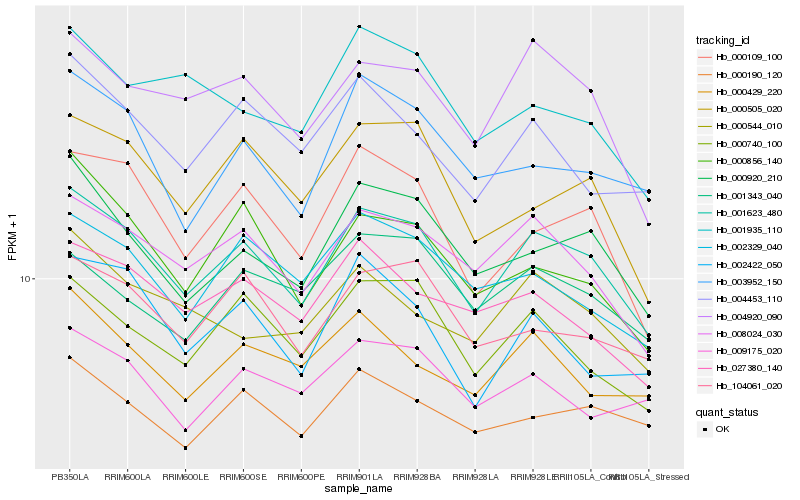
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_480 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 2 |
Hb_008024_030 |
0.0529640087 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 3 |
Hb_000920_210 |
0.0602912503 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 4 |
Hb_004920_090 |
0.0611125186 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000740_100 |
0.0628872548 |
- |
- |
calpain, putative [Ricinus communis] |
| 6 |
Hb_000109_100 |
0.0660392448 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 7 |
Hb_104061_020 |
0.0695370166 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 8 |
Hb_002329_040 |
0.071385783 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 9 |
Hb_027380_140 |
0.0723349439 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000505_020 |
0.0728188715 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 11 |
Hb_000190_120 |
0.0735021682 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 12 |
Hb_004453_110 |
0.0737064885 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 13 |
Hb_000429_220 |
0.0751718949 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003952_150 |
0.0752440712 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 15 |
Hb_009175_020 |
0.0766045605 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_000856_140 |
0.0770060836 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002422_050 |
0.077452089 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000544_010 |
0.0781453115 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_001343_040 |
0.0797085908 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 20 |
Hb_001935_110 |
0.0830350889 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |