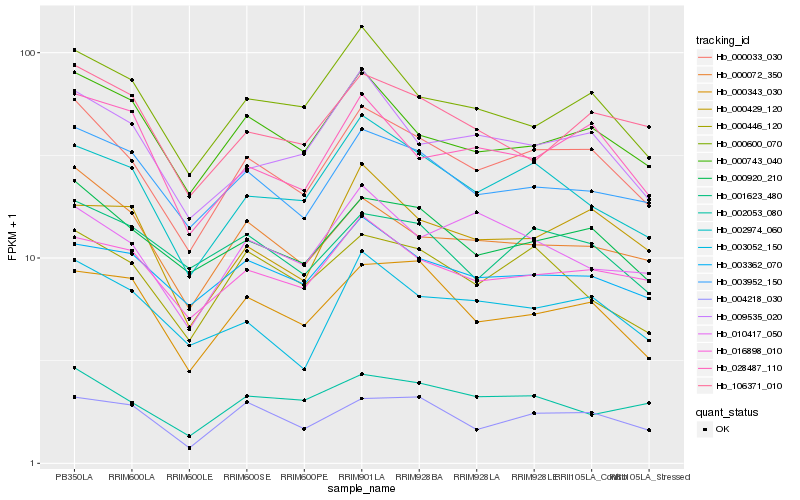
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000033_030 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 2 |
Hb_000920_210 |
0.0678416361 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 3 |
Hb_003952_150 |
0.0801923363 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 4 |
Hb_000072_350 |
0.0812679033 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 5 |
Hb_002974_060 |
0.083115641 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 6 |
Hb_000743_040 |
0.0841450002 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003052_150 |
0.0861315349 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 8 |
Hb_016898_010 |
0.0879862861 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 9 |
Hb_009535_020 |
0.0881436184 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 10 |
Hb_000600_070 |
0.0891906973 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 11 |
Hb_004218_030 |
0.0894139516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000446_120 |
0.0899309934 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 13 |
Hb_028487_110 |
0.0915797219 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 14 |
Hb_000343_030 |
0.0919665357 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 15 |
Hb_002053_080 |
0.0936346485 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21300 [Jatropha curcas] |
| 16 |
Hb_003362_070 |
0.0969287729 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 17 |
Hb_106371_010 |
0.0974734073 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 18 |
Hb_001623_480 |
0.097722894 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 19 |
Hb_000429_120 |
0.0982752211 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 20 |
Hb_010417_050 |
0.1002815918 |
- |
- |
PREDICTED: uncharacterized protein LOC105645746 [Jatropha curcas] |