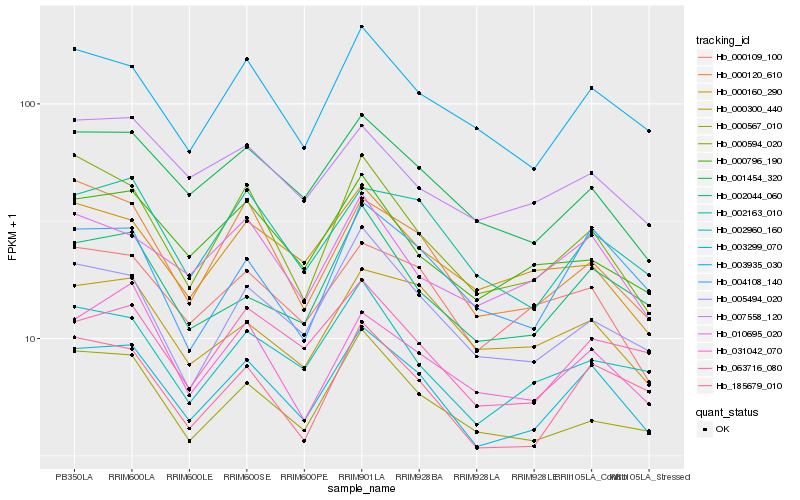
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003299_070 |
0.0 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 2 |
Hb_003935_030 |
0.0653981136 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 3 |
Hb_001454_320 |
0.0686610318 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_185679_010 |
0.0691362446 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 5 |
Hb_000109_100 |
0.0703944232 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 6 |
Hb_002163_010 |
0.0711923343 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 7 |
Hb_010695_020 |
0.0740784981 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 [Jatropha curcas] |
| 8 |
Hb_005494_020 |
0.0790560087 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 9 |
Hb_002044_060 |
0.0793472072 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 10 |
Hb_063716_080 |
0.0794737298 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 11 |
Hb_002960_160 |
0.0801007318 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 12 |
Hb_000796_190 |
0.0806760713 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 13 |
Hb_004108_140 |
0.0811997434 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000567_010 |
0.0812194706 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 15 |
Hb_000120_610 |
0.0812470618 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 16 |
Hb_031042_070 |
0.0820270001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007558_120 |
0.0828579206 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 18 |
Hb_000594_020 |
0.0829574374 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000300_440 |
0.0833014463 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000160_290 |
0.08466541 |
- |
- |
unnamed protein product [Vitis vinifera] |