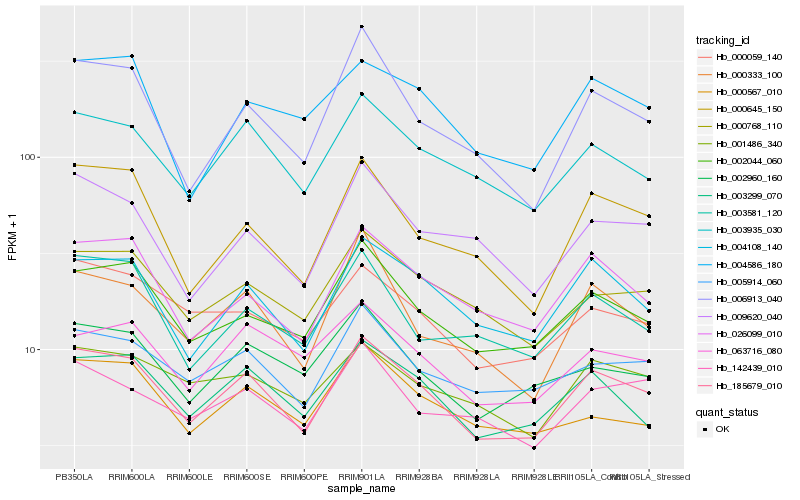
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185679_010 |
0.0 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 2 |
Hb_004108_140 |
0.0659749756 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_003935_030 |
0.0672516113 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 4 |
Hb_003299_070 |
0.0691362446 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 5 |
Hb_002044_060 |
0.0712070234 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 6 |
Hb_026099_010 |
0.0749571579 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 7 |
Hb_005914_060 |
0.075732562 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 8 |
Hb_000768_110 |
0.0781498813 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000059_140 |
0.0825338024 |
- |
- |
rnase h, putative [Ricinus communis] |
| 10 |
Hb_002960_160 |
0.0840284876 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 11 |
Hb_000645_150 |
0.0843899071 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_006913_040 |
0.086204978 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 13 |
Hb_063716_080 |
0.0883908518 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 14 |
Hb_001486_340 |
0.0888484798 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_000333_100 |
0.0898744436 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009620_040 |
0.0903281804 |
- |
- |
PREDICTED: cyclin-T1-3-like [Jatropha curcas] |
| 17 |
Hb_000567_010 |
0.0907236388 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 18 |
Hb_142439_010 |
0.090743469 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 19 |
Hb_004586_180 |
0.0907796979 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 20 |
Hb_003581_120 |
0.0911149628 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |