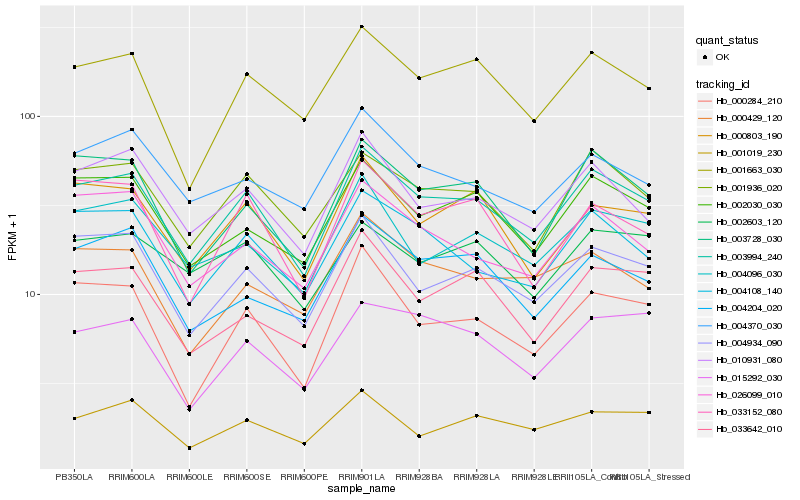
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_240 |
0.0 |
- |
- |
Mediator subunit 8 isoform 3 [Theobroma cacao] |
| 2 |
Hb_001936_020 |
0.0607548541 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 3 |
Hb_004934_090 |
0.0661272331 |
- |
- |
hypothetical protein JCGZ_08073 [Jatropha curcas] |
| 4 |
Hb_003728_030 |
0.0685527505 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 5 |
Hb_001663_030 |
0.0700966857 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 6 |
Hb_004108_140 |
0.0701412236 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 7 |
Hb_002030_030 |
0.0706854829 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 8 |
Hb_010931_080 |
0.0729811969 |
- |
- |
PREDICTED: U4/U6.U5 tri-snRNP-associated protein 2-like [Jatropha curcas] |
| 9 |
Hb_000803_190 |
0.0766090024 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004096_030 |
0.0818850192 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_033152_080 |
0.0841746748 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 12 |
Hb_026099_010 |
0.0842620525 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 13 |
Hb_000284_210 |
0.0862257077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004370_030 |
0.0865492878 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 15 |
Hb_001019_230 |
0.0874660858 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070 [Jatropha curcas] |
| 16 |
Hb_015292_030 |
0.0882786155 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02490, mitochondrial [Jatropha curcas] |
| 17 |
Hb_033642_010 |
0.0883243373 |
- |
- |
PREDICTED: protection of telomeres protein 1a-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004204_020 |
0.0884411642 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105640780 [Jatropha curcas] |
| 19 |
Hb_002603_120 |
0.0888673837 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 20 |
Hb_000429_120 |
0.0896743093 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |