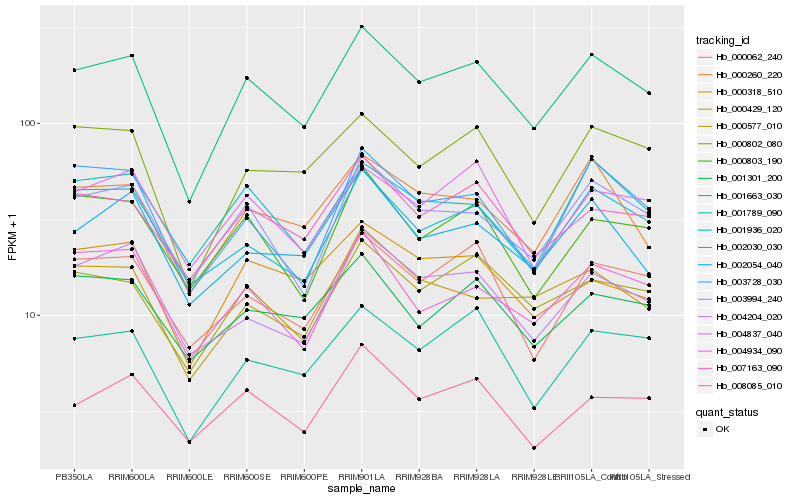
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001663_030 |
0.0 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 2 |
Hb_000577_010 |
0.0691174834 |
- |
- |
hypothetical protein JCGZ_23887 [Jatropha curcas] |
| 3 |
Hb_003994_240 |
0.0700966857 |
- |
- |
Mediator subunit 8 isoform 3 [Theobroma cacao] |
| 4 |
Hb_007163_090 |
0.0725348747 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 5 |
Hb_000802_080 |
0.0747893921 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 6 |
Hb_001789_090 |
0.0764027079 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |
| 7 |
Hb_000260_220 |
0.0776571519 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002030_030 |
0.0787536922 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 9 |
Hb_004837_040 |
0.0819386776 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 10 |
Hb_001936_020 |
0.0822693031 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 11 |
Hb_000062_240 |
0.0827285662 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 12 |
Hb_004934_090 |
0.0838665318 |
- |
- |
hypothetical protein JCGZ_08073 [Jatropha curcas] |
| 13 |
Hb_000429_120 |
0.0848475835 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 14 |
Hb_001301_200 |
0.0848751857 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 15 |
Hb_000318_510 |
0.0852590964 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 16 |
Hb_003728_030 |
0.0854706322 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 17 |
Hb_008085_010 |
0.0857220686 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g26680, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002054_040 |
0.0857413941 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 19 |
Hb_004204_020 |
0.0857963656 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105640780 [Jatropha curcas] |
| 20 |
Hb_000803_190 |
0.0864381505 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |