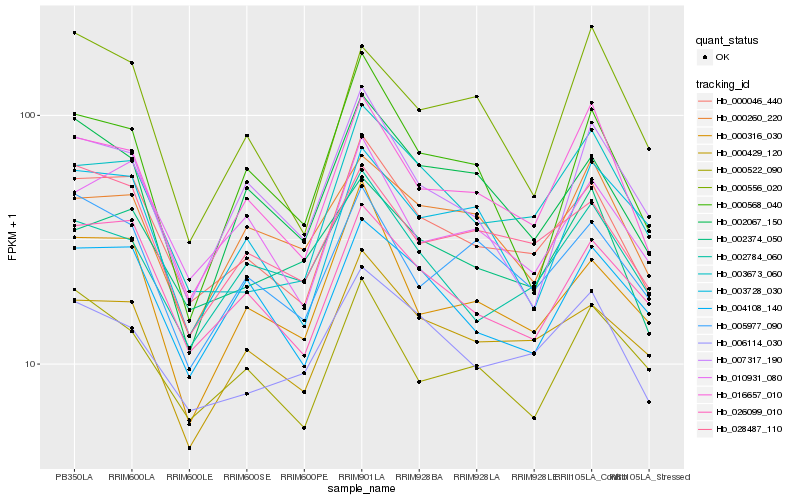
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016657_010 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 5 [Jatropha curcas] |
| 2 |
Hb_000046_440 |
0.0753317607 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 3 |
Hb_000260_220 |
0.0803399073 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007317_190 |
0.0890250306 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 5 |
Hb_000522_090 |
0.091038176 |
- |
- |
PREDICTED: actin-related protein 9 isoform X1 [Nicotiana tomentosiformis] |
| 6 |
Hb_005977_090 |
0.0913611038 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g16250 [Jatropha curcas] |
| 7 |
Hb_000556_020 |
0.0916092146 |
- |
- |
PREDICTED: terminal uridylyltransferase 7 [Jatropha curcas] |
| 8 |
Hb_002784_060 |
0.0939260464 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003728_030 |
0.0950608082 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 10 |
Hb_010931_080 |
0.0965204696 |
- |
- |
PREDICTED: U4/U6.U5 tri-snRNP-associated protein 2-like [Jatropha curcas] |
| 11 |
Hb_000568_040 |
0.096571638 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2 [Jatropha curcas] |
| 12 |
Hb_002374_050 |
0.0980230938 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 13 |
Hb_006114_030 |
0.0985352559 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 14 |
Hb_004108_140 |
0.0990969491 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000429_120 |
0.0992992911 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 16 |
Hb_028487_110 |
0.0993436185 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_026099_010 |
0.0995541271 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 18 |
Hb_002067_150 |
0.0997498554 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 19 |
Hb_003673_060 |
0.1007753145 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 20 |
Hb_000316_030 |
0.1018145158 |
- |
- |
zinc finger protein, putative [Ricinus communis] |