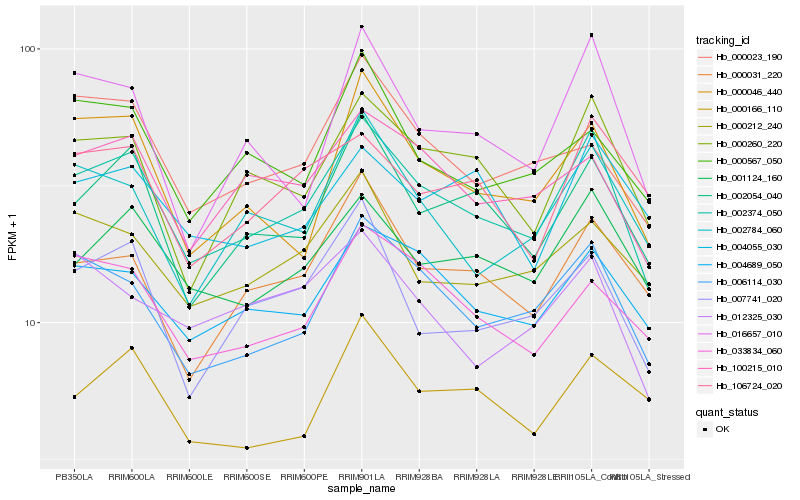
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_050 |
0.0 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 2 |
Hb_006114_030 |
0.0719764689 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 3 |
Hb_002054_040 |
0.0772498068 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 4 |
Hb_000260_220 |
0.0774684676 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004689_050 |
0.0788792123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001124_160 |
0.0793108349 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 7 |
Hb_100215_010 |
0.0832658312 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 8 |
Hb_000046_440 |
0.0901633301 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 9 |
Hb_007741_020 |
0.0907732454 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 10 |
Hb_002784_060 |
0.0912373655 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_033834_060 |
0.0916221733 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000031_220 |
0.0924997868 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 13 |
Hb_000212_240 |
0.0970539222 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000023_190 |
0.0975695583 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 15 |
Hb_016657_010 |
0.0980230938 |
- |
- |
PREDICTED: peroxisome biogenesis protein 5 [Jatropha curcas] |
| 16 |
Hb_106724_020 |
0.1004722005 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 1A-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000567_050 |
0.1006386554 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 18 |
Hb_004055_030 |
0.1030826336 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 19 |
Hb_000166_110 |
0.1032896037 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 25 [Jatropha curcas] |
| 20 |
Hb_012325_030 |
0.1033111794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |