| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
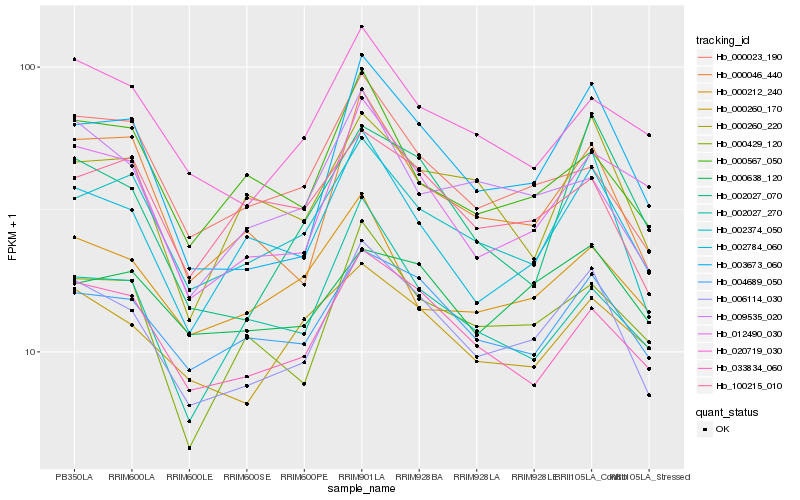
Hb_006114_030 |
0.0 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 2 |
Hb_002374_050 |
0.0719764689 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 3 |
Hb_004689_050 |
0.0727044528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033834_060 |
0.0768594629 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_003673_060 |
0.0770852442 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 6 |
Hb_000046_440 |
0.0787076942 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 7 |
Hb_002784_060 |
0.0788896041 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000023_190 |
0.0876763758 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 9 |
Hb_000429_120 |
0.0892659835 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 10 |
Hb_000260_170 |
0.0905527825 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 11 |
Hb_012490_030 |
0.0913534474 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_020719_030 |
0.0918815507 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 13 |
Hb_009535_020 |
0.0936673415 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 14 |
Hb_000212_240 |
0.0966982476 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000567_050 |
0.0969693384 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 16 |
Hb_000638_120 |
0.0969863441 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_002027_070 |
0.0970812355 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 18 |
Hb_002027_270 |
0.0976371487 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 19 |
Hb_100215_010 |
0.0979527289 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 20 |
Hb_000260_220 |
0.0984015588 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |