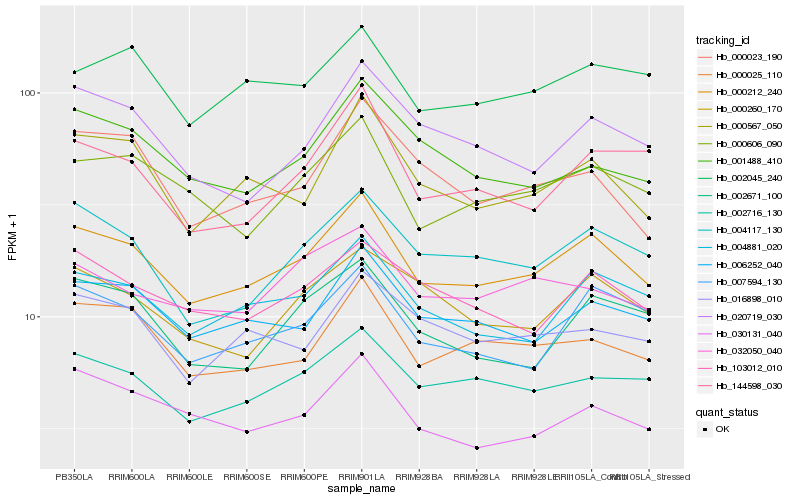
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_240 |
0.0 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_004881_020 |
0.0626674829 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 3 |
Hb_006252_040 |
0.0634544553 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002716_130 |
0.0670813381 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000025_110 |
0.0679954721 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 6 |
Hb_004117_130 |
0.0684493639 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 7 |
Hb_000606_090 |
0.0685679153 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 8 |
Hb_007594_130 |
0.0694223612 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000567_050 |
0.069779126 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 10 |
Hb_103012_010 |
0.0732541349 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 11 |
Hb_030131_040 |
0.0753747563 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 12 |
Hb_000260_170 |
0.0779868723 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 13 |
Hb_002045_240 |
0.0782842604 |
- |
- |
2-methyl-6-phytylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 14 |
Hb_002671_100 |
0.0784627477 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase-like [Jatropha curcas] |
| 15 |
Hb_020719_030 |
0.0784869004 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 16 |
Hb_144598_030 |
0.0812018314 |
- |
- |
PREDICTED: mitotic apparatus protein p62 [Jatropha curcas] |
| 17 |
Hb_016898_010 |
0.0815702826 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 18 |
Hb_000023_190 |
0.082115022 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 19 |
Hb_032050_040 |
0.0828176222 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 20 |
Hb_001488_410 |
0.0828659983 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |