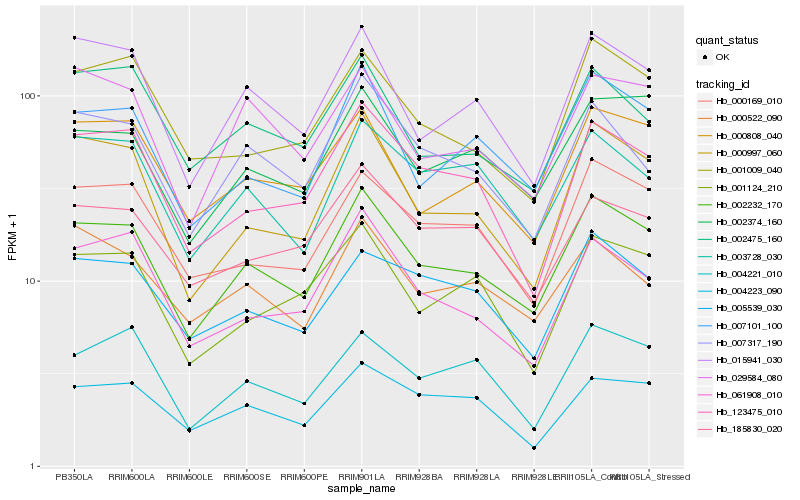
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 2 |
Hb_007101_100 |
0.0750619881 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 3 |
Hb_015941_030 |
0.0753009861 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_000997_060 |
0.0763572437 |
- |
- |
PREDICTED: DNA repair protein XRCC4 [Jatropha curcas] |
| 5 |
Hb_000169_010 |
0.0796621551 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_002374_160 |
0.0808833351 |
- |
- |
PREDICTED: N-glycosylase/DNA lyase OGG1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000808_040 |
0.0827615158 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 3 [Jatropha curcas] |
| 8 |
Hb_123475_010 |
0.0843296789 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 9 |
Hb_007317_190 |
0.0876276701 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 10 |
Hb_003728_030 |
0.088227967 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 11 |
Hb_005539_030 |
0.0896610911 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_029584_080 |
0.0904451765 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 13 |
Hb_001009_040 |
0.0922211735 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Jatropha curcas] |
| 14 |
Hb_061908_010 |
0.0926380542 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 15 |
Hb_001124_210 |
0.0928860486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_185830_020 |
0.0933792322 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_002475_160 |
0.0938095947 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 18 |
Hb_004223_090 |
0.0951451822 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X2 [Jatropha curcas] |
| 19 |
Hb_004221_010 |
0.095443855 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 20 |
Hb_000522_090 |
0.0959971505 |
- |
- |
PREDICTED: actin-related protein 9 isoform X1 [Nicotiana tomentosiformis] |