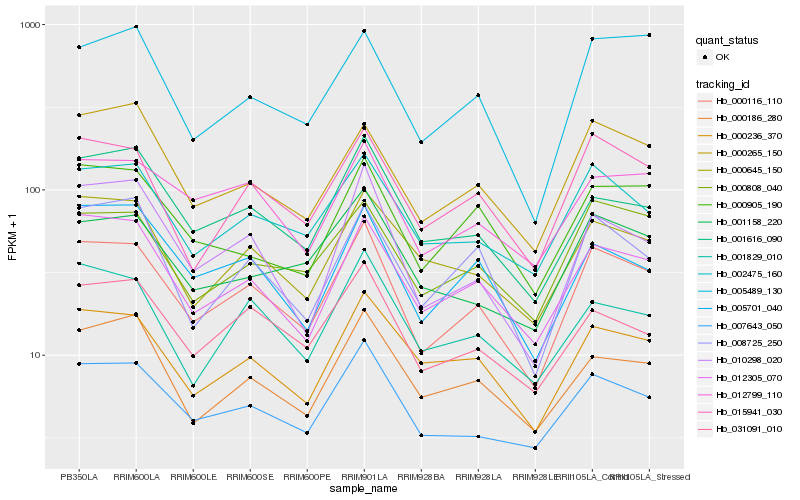
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_110 |
0.0 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 2 |
Hb_007643_050 |
0.0725387812 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_012305_070 |
0.0766145833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015941_030 |
0.0776881825 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_002475_160 |
0.0815831083 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 6 |
Hb_000236_370 |
0.0856833252 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 7 |
Hb_000265_150 |
0.0875181281 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012799_110 |
0.0887085998 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 9 |
Hb_000808_040 |
0.0890208154 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 3 [Jatropha curcas] |
| 10 |
Hb_031091_010 |
0.0891433497 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 11 |
Hb_001616_090 |
0.0913805472 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_010298_020 |
0.0924167071 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 isoform X4 [Vitis vinifera] |
| 13 |
Hb_005701_040 |
0.0937363836 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_008725_250 |
0.0937806709 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000645_150 |
0.094475366 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000186_280 |
0.0951976394 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_005489_130 |
0.0954757088 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 18 |
Hb_001829_010 |
0.0958228151 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000905_190 |
0.0972984294 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 20 |
Hb_001158_220 |
0.0985034655 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |